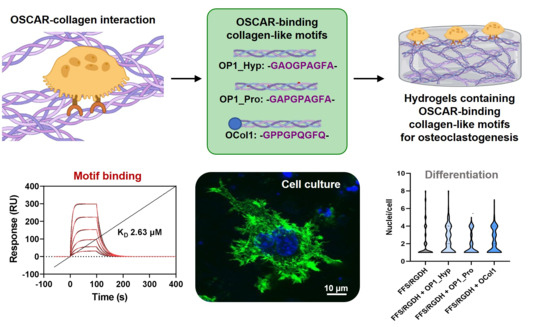
Collagen-like Osteoclast-Associated Receptor (OSCAR)-Binding Motifs Show a Co-Stimulatory Effect on Osteoclastogenesis in a Peptide Hydrogel System
Abstract
:1. Introduction
2. Results and Discussion
2.1. OP1_Hyp, OP1_Pro, and OCol1 Adopt a Collagen Triple Helical Structure
2.2. Both OP1_Hyp and OP1_Pro Bind to OSCAR, but with Different Affinities
2.3. OSCAR Contributes to Raw 264.7 Cell Differentiation towards OCs
2.4. OSCAR-Modified Hydrogels for OC Culture and Differentiation
3. Materials and Methods
3.1. OSCAR-Binding Peptides and Recombinant Protein
3.2. Circular Dichroism (CD) Analysis of Secondary Structure and Thermal Stability
3.3. Surface Plasmon Resonance (SPR) Analysis of the Peptide-OSCAR Interaction
3.4. Raw 264.7 Cell Adhesion Assay
3.5. Peptides and Peptide Hydrogel Fabrication
3.6. Cell Culture and Assessment of Viability and Morphology
3.7. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
3.8. Gene Expression
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyce, B.F.; Xing, L. The RANKL/RANK/OPG pathway. Curr. Osteoporos. Rep. 2007, 5, 98–104. [Google Scholar] [CrossRef]
- Takegahara, N.; Kim, H.; Choi, Y. RANKL biology. Bone 2022, 159, 116353. [Google Scholar] [CrossRef]
- Elson, A.; Anuj, A.; Barnea-Zohar, M.; Reuven, N. The origins and formation of bone-resorbing osteoclasts. Bone 2022, 164, 116538. [Google Scholar] [CrossRef]
- Nedeva, I.R.; Vitale, M.; Elson, A.; Hoyland, J.A.; Bella, J. Role of OSCAR Signaling in Osteoclastogenesis and Bone Disease. Front. Cell Dev. Biol. 2021, 9, 641162. [Google Scholar] [CrossRef]
- Barrow, A.D.; Raynal, N.; Andersen, T.L.; Slatter, D.A.; Bihan, D.; Pugh, N.; Cella, M.; Kim, T.; Rho, J.; Negishi-Koga, T.; et al. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J. Clin. Investig. 2011, 121, 3505–3516. [Google Scholar] [CrossRef]
- Jansen, S.M.; Willumsen, N.; Karsdal, M.A. Collagen receptors. In Biochemistry of Collagens, Laminins and Elastin, 3rd ed.; Karsdal, M.A., Ed.; Academic Press: Cambridge, MA, USA, 2024; pp. 317–336. ISBN 978-0443156175. [Google Scholar]
- Kim, G.M.; Park, H.; Lee, S.Y. Roles of osteoclast-associated receptor in rheumatoid arthritis and osteoarthritis. Jt. Bone Spine 2022, 89, 105400. [Google Scholar] [CrossRef]
- Kim, N.; Takami, M.; Rho, J.; Josien, R.; Choi, Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J. Exp. Med. 2002, 195, 201–209. [Google Scholar] [CrossRef]
- Zhou, L.; Hinerman, J.M.; Blaszczyk, M.; Miller, J.L.C.; Conrady, D.G.; Barrow, A.D.; Chirgadze, D.Y.; Bihan, D.; Farndale, R.W.; Herr, A.B. Structural basis for collagen recognition by the immune receptor OSCAR. Blood 2016, 127, 529–537. [Google Scholar] [CrossRef]
- Schultz, H.S.; Guo, L.; Keller, P.; Fleetwood, A.J.; Sun, M.; Guo, W.; Ma, C.; Hamilton, J.A.; Bjørkdahl, O.; Berchtold, M.W.; et al. OSCAR-collagen signaling in monocytes plays a proinflammatory role and may contribute to the pathogenesis of rheumatoid arthritis. Eur. J. Immunol. 2016, 46, 952–963. [Google Scholar] [CrossRef]
- Liao, X.; Bu, Y.; Zhang, Y.; Xu, B.; Liang, J.; Jia, Q.; Zhang, C. OSCAR facilitates malignancy with enhanced metastasis correlating to inhibitory immune microenvironment in multiple cancer types. J. Cancer 2021, 12, 3769–3780. [Google Scholar] [CrossRef]
- Bella, J. Collagen structure: New tricks from a very old dog. Biochem. J. 2016, 473, 1001–1025. [Google Scholar] [CrossRef] [PubMed]
- Haywood, J.; Qi, J.; Chen, C.-C.; Lu, G.; Liu, Y.; Yan, J.; Shi, Y.; Gao, G.F. Structural basis of collagen recognition by human osteoclast-associated receptor and design of osteoclastogenesis inhibitors. Proc. Natl. Acad. Sci. USA 2016, 113, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xing, R.; Bai, S.; Yan, X. Recent advances of self-assembling peptide-based hydrogels for biomedical applications. Soft Matter 2019, 15, 1704–1715. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; He, H.; Li, B.; Hou, T. Hydrogel as a Biomaterial for Bone Tissue Engineering: A Review. Nanomaterials 2020, 10, 1511. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Song, J.; Zhang, X.; Jiang, K.; Fan, H.; Li, Y.; Zhao, Y.; Liu, S.; Hao, D.; Li, G. Hydroxyapatite-tethered peptide hydrogel promotes osteogenesis. Gels 2022, 8, 804. [Google Scholar] [CrossRef] [PubMed]
- Alshehri, S.; Susapto, H.H.; Hauser, C.A.E. Scaffolds from self-assembling tetrapeptides support 3D spreading, osteogenic differentiation, and angiogenesis of mesenchymal stem cells. Biomacromolecules 2021, 22, 2094–2106. [Google Scholar] [CrossRef] [PubMed]
- Ligorio, C.; Mata, A. Synthetic extracellular matrices with function-encoding peptides. Nat. Rev. Bioeng. 2023, 1, 518–536. [Google Scholar] [CrossRef]
- Malcor, J.D.; Mallein-Gerin, F. Biomaterial functionalization with triple-helical peptides for tissue engineering. Acta Biomater. 2022, 148, 1–21. [Google Scholar] [CrossRef]
- Vitale, M.; Ligorio, C.; McAvan, B.; Hodson, N.W.; Allan, C.; Richardson, S.M.; Hoyland, J.A.; Bella, J. Hydroxyapatite-decorated Fmoc-hydrogel as a bone-mimicking substrate for osteoclast differentiation and culture. Acta Biomater. 2022, 138, 144–154. [Google Scholar] [CrossRef]
- Vitale, M.; Ligorio, C.; Smith, I.P.; Richardson, S.M.; Hoyland, J.A.; Bella, J. Incorporation of Natural and Recombinant Collagen Proteins within Fmoc-Based Self-Assembling Peptide Hydrogels. Gels 2022, 8, 254. [Google Scholar] [CrossRef]
- Ghosh, N.; McKillop, T.J.; Jowitt, T.A.; Howard, M.; Davies, H.; Holmes, D.F.; Roberts, I.S.; Bella, J. Collagen-Like Proteins in Pathogenic E. coli Strains. PLoS ONE 2012, 7, e37872. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 2006, 1, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
- Drzewiecki, K.E.; Grisham, D.R.; Parmar, A.S.; Nanda, V.; Shreiber, D.I. Circular Dichroism Spectroscopy of Collagen Fibrillogenesis: A New Use for an Old Technique. Biophys. J. 2016, 111, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.; Price, N. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef]
- Jönsson, U.; Fägerstam, L.; Ivarsson, B.; Johnsson, B.; Karlsson, R.; Lundh, K.; Löfås, S.; Persson, B.; Roos, H.; Rönnberg, I. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. Biotechniques 1991, 11, 620–627. [Google Scholar]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef]
- Song, C.; Yang, X.; Lei, Y.; Zhang, Z.; Smith, W.; Yan, J.; Kong, L. Evaluation of efficacy on RANKL induced osteoclast from RAW264.7 cells. J. Cell. Physiol. 2019, 234, 11969–11975. [Google Scholar] [CrossRef]
- Kodama, J.; Kaito, T. Osteoclast multinucleation: Review of current literature. Int. J. Mol. Sci. 2020, 21, 5685. [Google Scholar] [CrossRef]
- Roscher, A.; Hasegawa, T.; Dohnke, S.; Ocaña-Morgner, C.; Amizuka, N.; Jessberger, R.; Garbe, A.I. The F-actin modulator SWAP-70 controls podosome patterning in osteoclasts. Bone Rep. 2016, 5, 214–221. [Google Scholar] [CrossRef]
- Lakkakorpi, P.T.; Väänänen, K.H. Kinetics of the osteoclast cytoskeleton during the resorption cycle in vitro. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 1991, 6, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Pierce, A.M.; Lindskog, S.; Hammarström, L. Osteoclasts: Structure and function. Electron Microsc. Rev. 1991, 4, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Lampiasi, N.; Russo, R.; Kireev, I.; Strelkova, O.; Zhironkina, O.; Zito, F. Osteoclasts Differentiation from Murine RAW 264.7 Cells Stimulated by RANKL: Timing and Behavior. Biology 2021, 10, 117. [Google Scholar] [CrossRef]








| Molecule | Sequence | Amino Acids | Monomer Mw (Da) | Trimer Mw (Da) |
|---|---|---|---|---|
| OP1_Hyp | Ac-GPOGPOGPOGAOGPAGFAGPOGPOGPO-NH2 | 27 | 2404.56 | 7213.68 |
| OP1_Pro | Ac-GPOGPOGPOGAPGPAGFAGPOGPOGPO-NH2 | 27 | 2388.56 | 7165.68 |
| OCol1 | MGSHHHHHHSGLVPRGSGPPGPPGPQGPAGPRGEPGPAGPKGEPGPAGPPGPQGFQGPPGPQGPAGPIGPKGEPGPIGPQGPKGDPGETQIRFRLGPASIIETNSNGWFPDTDGALITGLTFLAPKDATRVQGFFQHLQVRFGDGPWQDVKGLDEVGSDTGRTGE | 165 | 16,625.34 | 49,876.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, M.; Ligorio, C.; Richardson, S.M.; Hoyland, J.A.; Bella, J. Collagen-like Osteoclast-Associated Receptor (OSCAR)-Binding Motifs Show a Co-Stimulatory Effect on Osteoclastogenesis in a Peptide Hydrogel System. Int. J. Mol. Sci. 2024, 25, 445. https://doi.org/10.3390/ijms25010445
Vitale M, Ligorio C, Richardson SM, Hoyland JA, Bella J. Collagen-like Osteoclast-Associated Receptor (OSCAR)-Binding Motifs Show a Co-Stimulatory Effect on Osteoclastogenesis in a Peptide Hydrogel System. International Journal of Molecular Sciences. 2024; 25(1):445. https://doi.org/10.3390/ijms25010445
Chicago/Turabian StyleVitale, Mattia, Cosimo Ligorio, Stephen M. Richardson, Judith A. Hoyland, and Jordi Bella. 2024. "Collagen-like Osteoclast-Associated Receptor (OSCAR)-Binding Motifs Show a Co-Stimulatory Effect on Osteoclastogenesis in a Peptide Hydrogel System" International Journal of Molecular Sciences 25, no. 1: 445. https://doi.org/10.3390/ijms25010445