Genome-Wide Identification and Expression Pattern Analysis of Dirigent Members in the Genus Oryza
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogeny Analysis of DIR Proteins
2.2. Gene Location and Duplication Events of DIR Proteins
2.3. Synteny Analysis of DIR Genes in the Genus Oryza
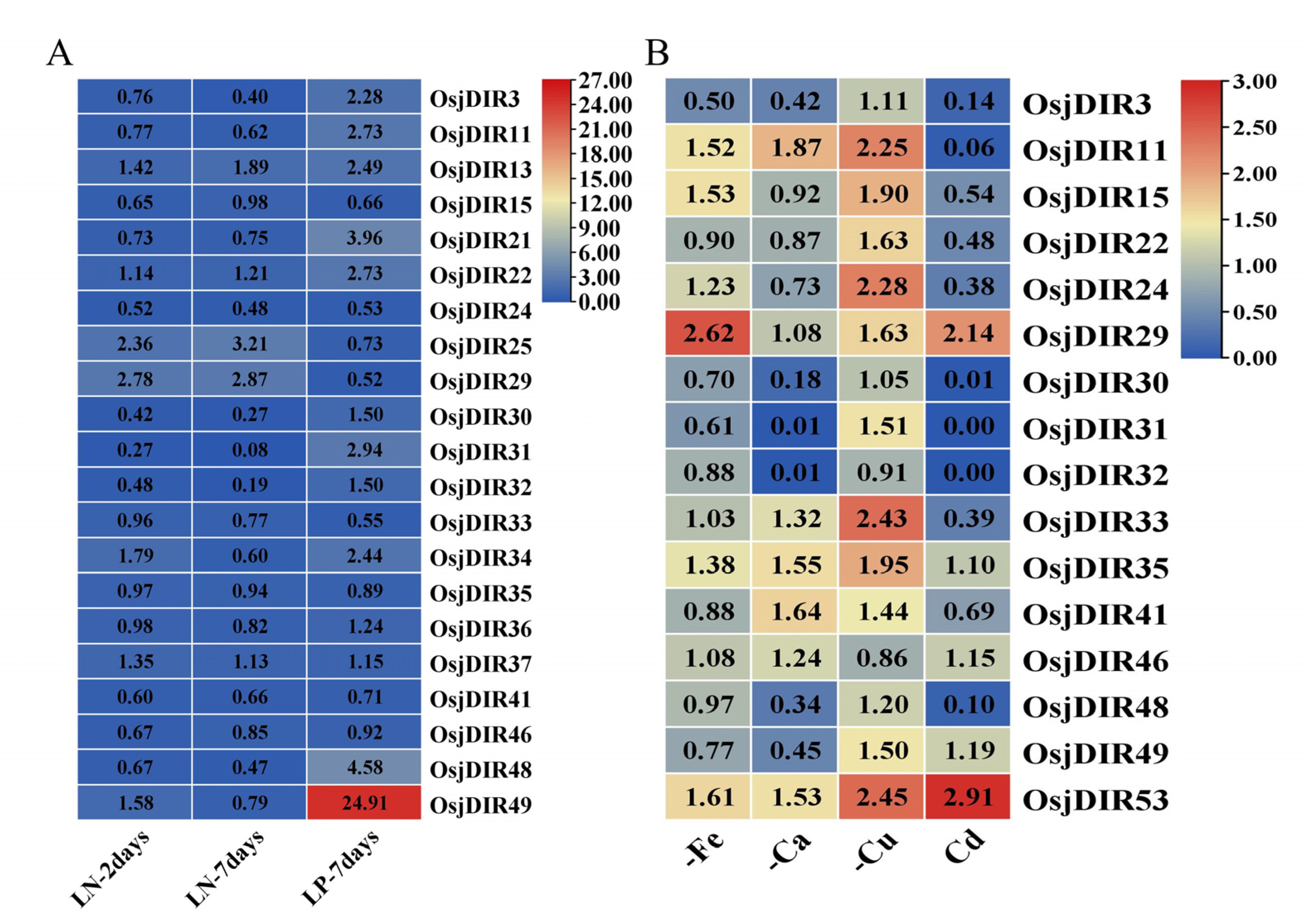
2.4. Bioinformatics Analysis of the Expression Patterns of OsjDIR Genes
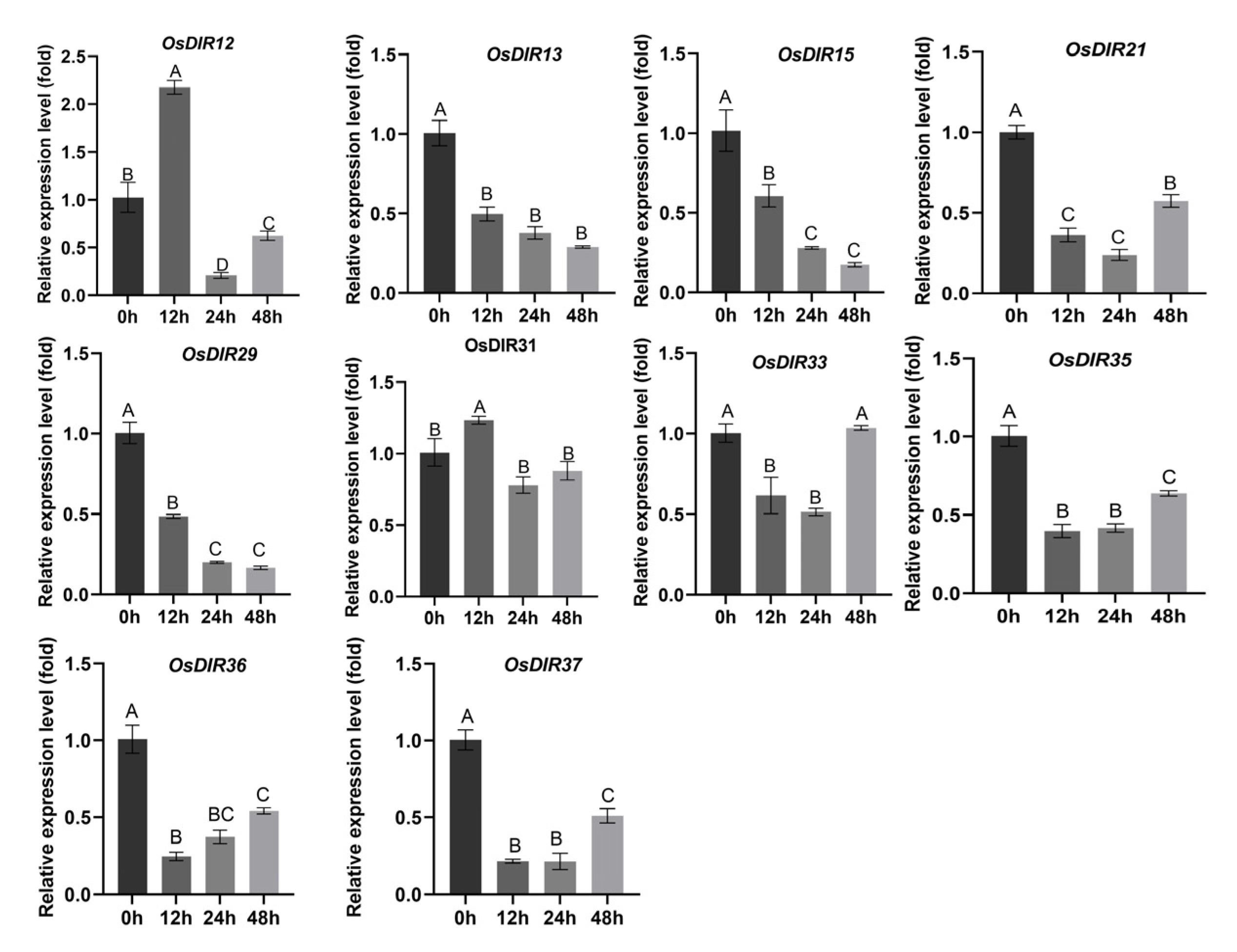
2.5. Experimental Examinations of the Expression Dynamics of OsjDIR Genes
2.6. Expression of OsjDIR Genes to Rhizoctonia solani Inoculation
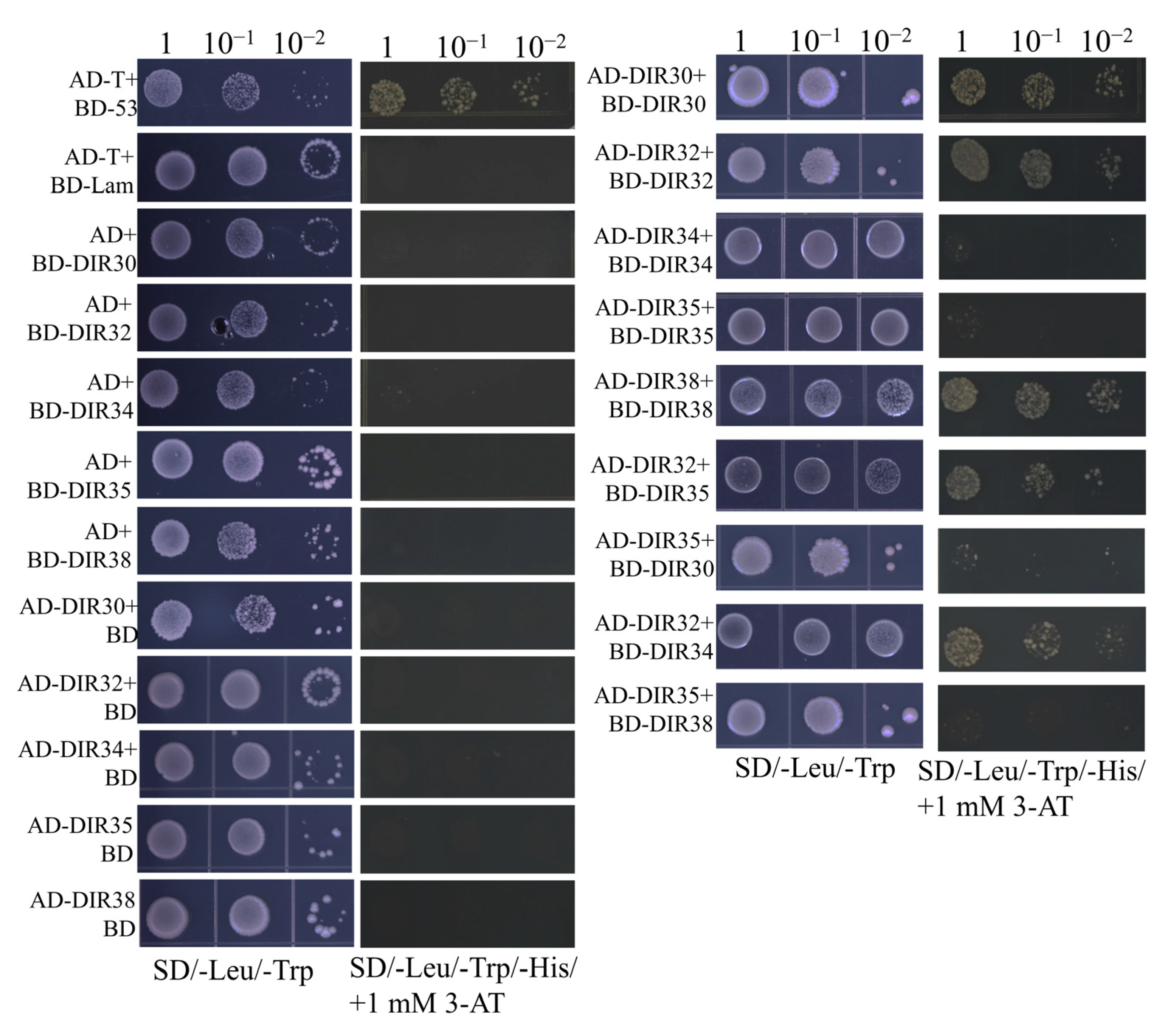
2.7. Many DIR Proteins Interact with Themselves or Other DIR Proteins
3. Discussion
4. Materials and Methods
4.1. Identification of DIR Genes in the Rice Species
4.2. Phylogenetic Analysis of DIR Proteins in the Different Rice Species
4.3. Gene Duplication Analysis and the Calculation of Ka/Ks Ratios
4.4. Expression Patterns of Rice DIR Genes through Analyzing the RNA-Seq Data
4.5. Plant Materials and Growth Conditions
4.6. Plant Materials’ Treatments
4.7. RNA Extraction and qRT-PCR Analysis
4.8. Yeast Two-Hybrid Assays
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davin, L.B.; Wang, H.B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective Bimolecular Phenoxy Radical Coupling by an Auxiliary (Dirigent) Protein without an Active Center. Science 1997, 275, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Bilkova, A.; Jackson, P.; Dabravolski, S.; Riber, W.; Didi, V.; Houser, J.; Gigli-Bisceglia, N.; Wimmerova, M.; Budínská, E.; et al. Dirigent Proteins in Plants: Modulating Cell Wall Metabolism during Abiotic and Biotic Stress Exposure. J. Exp. Bot. 2017, 68, 3287–3301. [Google Scholar] [CrossRef]
- Khan, A.; Li, R.J.; Sun, J.T.; Ma, F.; Zhang, H.X.; Jin, J.H.; Ali, M.; Haq, S.U.; Wang, J.E.; Gong, Z.H. Genome-Wide Analysis of Dirigent Gene Family in Pepper (Capsicum annuum L.) and Characterization of CaDIR7 in Biotic and Abiotic Stresses. Sci. Rep. 2018, 8, 5500. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.W.; Wang, X.F.; Sun, Z.W.; Zhang, Y.; Meng, C.S.; Chen, B.; Wang, G.N.; Ke, H.F.; Wu, J.H.; Yan, Y.Y.; et al. Evolution, Expression and Functional Analysis of Cultivated Allotetraploid Cotton DIR Genes. BMC Plant Biol. 2021, 21, 89. [Google Scholar] [CrossRef]
- Dalisay, D.S.; Kim, K.W.; Lee, C.; Yang, H.; Rübel, O.; Bowen, B.P.; Davin, L.B.; Lewis, N.G. Dirigent Protein-Mediated Lignan and Cyanogenic Glucoside Formation in Flax Seed: Integrated Omics and MALDI Mass Spectrometry Imaging. J. Nat. Prod. 2015, 78, 1231–1242. [Google Scholar] [CrossRef]
- Xu, W.Y.; Liu, T.; Zhang, H.Y.; Zhu, H. Mungbean DIRIGENT Gene Subfamilies and Their Expression Profiles Under Salt and Drought Stresses. Front. Genet. 2021, 12, 658148. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.F.; Xiao, Y.; Di, P.; Zhang, L.; Chen, W.S. The Dirigent Multigene Family in Isatis indigotica: Gene Discovery and Differential Transcript Abundance. BMC Genom. 2014, 15, 388. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Koyama, T.; Bahabadi, S.E.; Matsumoto, E.; Ono, E.; Murata, J. Essences in Metabolic Engineering of Lignan Biosynthesis. Metabolites 2015, 5, 270–290. [Google Scholar] [CrossRef]
- Davin, L.B.; Jourdes, M.; Patten, A.M.; Kim, K.W.; Vassão, D.G.; Lewis, N.G. Dissection of Lignin Macromolecular Configuration and Assembly: Comparison to Related Biochemical Processes in Allyl/Propenyl Phenol and Lignan Biosynthesis. Nat. Prod. Rep. 2008, 25, 1015–1090. [Google Scholar] [CrossRef] [PubMed]
- Davin, L.B.; Lewis, N.G. Lignin Primary Structures and Dirigent Sites. Curr. Opin. Biotechnol. 2005, 16, 407–415. [Google Scholar] [CrossRef]
- Li, N.H.; Zhao, M.; Liu, T.F.; Dong, L.D.; Cheng, Q.; Wu, J.J.; Wang, L.; Chen, X.; Zhang, C.Z.; Lu, W.C.; et al. A Novel Soybean Dirigent Gene GmDIR22 Contributes to Promotion of Lignan Biosynthesis and Enhances Resistance to Phytophthora sojae. Front. Plant Sci. 2017, 8, 1185. [Google Scholar] [CrossRef]
- Shi, H.Y.; Liu, Z.H.; Zhu, L.; Zhang, C.J.; Chen, Y.; Zhou, Y.; Li, F.G.; Li, X.B. Overexpression of Cotton (Gossypium hirsutum) Dirigent 1 Gene Enhances Lignification That Blocks the Spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhong, S.B.; Zhang, Q.J.; Ren, Y.; Sun, C.W.; Chen, F. A Loss-of-Function of the Dirigent Gene TaDIR-B1 Improves Resistance to Fusarium crown rot in Wheat. Plant Biotechnol. J. 2021, 19, 866–868. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yan, W.; Sun, L.; Tian, J.; Liao, H. Proteomic Analysis Reveals Growth Inhibition of Soybean Roots by Manganese Toxicity Is Associated with Alteration of Cell Wall Structure and Lignification. J. Proteom. 2016, 143, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.H.; Wang, L.; Wang, Z.; Shang, H.H.; Liu, X.; Zhu, Y.; Qi, D.D.; Deng, X. Cloning and Expression Analysis of a Dirigent Protein Gene from the Resurrection Plant Boea hygrometrica. Prog. Nat. Sci. 2009, 19, 347–352. [Google Scholar] [CrossRef]
- Gao, C.Q.; Liu, G.F.; Wang, Y.C.; Jing, J.; Yang, C.P. Cloning and analysis of dirigent-like protein in gene from Tamarix androssowii. Bull. Bot. Res. 2010, 30, 81–86. [Google Scholar]
- Guo, J.L.; Xu, L.P.; Fang, J.P.; Su, Y.C.; Fu, H.Y.; Que, Y.X.; Xu, J.S. A Novel Dirigent Protein Gene with Highly Stem-Specific Expression from Sugarcane, Response to Drought, Salt and Oxidative Stresses. Plant Cell Rep. 2012, 31, 1801–1812. [Google Scholar]
- Hosmani, P.S.; Kamiya, T.; Danku, J.; Naseer, S.; Geldner, N.; Guerinot, M.L.; Salt, D.E. Dirigent Domain-Containing Protein Is Part of the Machinery Required for Formation of the Lignin-Based Casparian Strip in the Root. Proc. Natl. Acad. Sci. USA 2013, 110, 14498–14503. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Cao, Y.B.; Liang, X.Y.; Zhuang, J.H.; Wang, X.F.; Qin, F.; Jiang, C.F. A Dirigent Family Protein Confers Variation of Casparian Strip Thickness and Salt Tolerance in Maize. Nat. Commun. 2022, 13, 2222. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Ranathunge, K.; Nayak, S.; Schreiber, L.; Mathew, M.K. Root Apoplastic Barriers Block Na+ Transport to Shoots in Rice (Oryza sativa L.). J. Exp. Bot. 2011, 62, 4215–4228. [Google Scholar] [CrossRef]
- Geldner, N. Casparian Strips. Curr. Biol. 2013, 23, R1025–R1026. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Schreiber, L.; Franke, R.B.; Geldner, N.; Reina-Pinto, J.J.; Kunst, L. Apoplastic Diffusion Barriers in Arabidopsis. Arab. Book Am. Soc. Plant Biol. 2013, 11, e0167. [Google Scholar] [CrossRef]
- Robbins, N.E.; Trontin, C.; Duan, L.; Dinneny, J.R. Beyond the Barrier: Communication in the Root through the Endodermis. Plant Physiol. 2014, 166, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Namyslov, J.; Bauriedlová, Z.; Janoušková, J.; Soukup, A.; Tylová, E. Exodermis and Endodermis Respond to Nutrient Deficiency in Nutrient-Specific and Localized Manner. Plants Basel Switz. 2020, 9, 201. [Google Scholar] [CrossRef] [PubMed]
- Armand, T.; Cullen, M.; Boiziot, F.; Li, L.; Fricke, W. Cortex Cell Hydraulic Conductivity, Endodermal Apoplastic Barriers and Root Hydraulics Change in Barley (Hordeum vulgare L.) in Response to a Low Supply of N and P. Ann. Bot. 2019, 124, 1091–1107. [Google Scholar] [CrossRef]
- Ge, S.; Sang, T.; Lu, B.R.; Hong, D.Y. Phylogeny of Rice Genomes with Emphasis on Origins of Allotetraploid Species. Proc. Natl. Acad. Sci. USA 1999, 96, 14400–14405. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Zheng, X.; Luo, J.; Gaut, B.S.; Ge, S. Multilocus Analysis of Nucleotide Variation of Oryza Sativa and Its Wild Relatives: Severe Bottleneck during Domestication of Rice. Mol. Biol. Evol. 2007, 24, 875–888. [Google Scholar] [CrossRef]
- Ralph, S.G.; Jancsik, S.; Bohlmann, J. Dirigent Proteins in Conifer Defense II: Extended Gene Discovery, Phylogeny, and Constitutive and Stress-Induced Gene Expression in Spruce (Picea Spp.). Phytochemistry 2007, 68, 1975–1991. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A Novel Insight into Functional Divergence of the MST Gene Family in Rice Based on Comprehensive Expression Patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef]
- Andreasson, E.; Jenkins, T.; Brodersen, P.; Thorgrimsen, S.; Petersen, N.H.T.; Zhu, S.; Qiu, J.L.; Micheelsen, P.; Rocher, A.; Petersen, M.; et al. The MAP Kinase Substrate MKS1 Is a Regulator of Plant Defense Responses. EMBO J. 2005, 24, 2579–2589. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Takeda, M.; Kidokoro, S.; Yamada, K.; Sakuma, Y.; Urano, K.; Fujita, M.; Yoshiwara, K.; Matsukura, S.; Morishita, Y.; et al. Metabolic Pathways Involved in Cold Acclimation Identified by Integrated Analysis of Metabolites and Transcripts Regulated by DREB1A and DREB2A. Plant Physiol. 2009, 150, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Abeysinghe, J.K.; Lam, K.M.; Ng, D.W.K. Differential Regulation and Interaction of Homoeologous WRKY18 and WRKY40 in Arabidopsis Allotetraploids and Biotic Stress Responses. Plant J. Cell Mol. Biol. 2019, 97, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Y.; Zhang, W.W.; Qu, H.Y.; Gou, S.S.; Li, L.X.; Song, H.H.; Yang, H.Q.; Li, W.J.; Zhang, H.; Hu, K.D.; et al. Transcriptomics Reveals the ERF2-BHLH2-CML5 Module Responses to H2S and ROS in Postharvest Calcium Deficiency Apples. Int. J. Mol. Sci. 2021, 22, 13013. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zeng, Y.N.; Yin, Y.B.; Pu, Y.Q.; Jackson, L.A.; Engle, N.L.; Martin, M.Z.; Tschaplinski, T.J.; Ding, S.Y.; Ragauskas, A.J.; et al. Pinoresinol Reductase 1 Impacts Lignin Distribution during Secondary Cell Wall Biosynthesis in Arabidopsis. Phytochemistry 2015, 112, 170–178. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Lee, C.H.; Zhou, J.L.; McCarthy, R.L.; Ye, Z.H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the Informatics Resource for Rice Transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef]
- Li, N.; Lin, B.; Wang, H.; Li, X.M.; Yang, F.F.; Ding, X.H.; Yan, J.B.; Chu, Z.H. Natural Variation in ZmFBL41 Confers Banded Leaf and Sheath Blight Resistance in Maize. Nat. Genet. 2019, 51, 1540–1548. [Google Scholar] [CrossRef]
- Cao, W.L.; Zhang, H.M.; Zhou, Y.; Zhao, J.H.; Lu, S.B.; Wang, X.Q.; Chen, X.J.; Yuan, L.M.; Guan, H.Y.; Wang, G.D.; et al. Suppressing Chlorophyll Degradation by Silencing OsNYC3 Improves Rice Resistance to Rhizoctonia solani, the Causal Agent of Sheath Blight. Plant Biotechnol. J. 2022, 20, 335–349. [Google Scholar] [CrossRef]
- Kim, K.W.; Smith, C.A.; Daily, M.D.; Cort, J.R.; Davin, L.B.; Lewis, N.G. Trimeric Structure of (+)-Pinoresinol-Forming Dirigent Protein at 1.95 Å Resolution with Three Isolated Active Sites. J. Biol. Chem. 2015, 290, 1308–1318. [Google Scholar] [CrossRef]
- Gasper, R.; Effenberger, I.; Kolesinski, P.; Terlecka, B.; Hofmann, E.; Schaller, A. Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6. Plant Physiol. 2016, 172, 2165–2175. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Huang, Q.F.; Gao, D.Y.; Wang, J.Y.; Lang, Y.S.; Liu, T.Y.; Li, B.; Bai, Z.T.; Luis, G.J.; Liang, C.Z.; et al. Whole-Genome Sequencing of Oryza Brachyantha Reveals Mechanisms Underlying Oryza Genome Evolution. Nat. Commun. 2013, 4, 1595. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Lainé, É.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A Genome-Wide Analysis of the Flax (Linum usitatissimum L.) Dirigent Protein Family: From Gene Identification and Evolution to Differential Regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Zou, C.; Lehti-Shiu, M.D.; Shinozaki, K.; Shiu, S.-H. Importance of Lineage-Specific Expansion of Plant Tandem Duplicates in the Adaptive Response to Environmental Stimuli. Plant Physiol. 2008, 148, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.; Hosmani, P.S.; Rus, A.; Lahner, B.; Borevitz, J.O.; Muthukumar, B.; Mickelbart, M.V.; Schreiber, L.; Franke, R.B.; Salt, D.E. Root Suberin Forms an Extracellular Barrier That Affects Water Relations and Mineral Nutrition in Arabidopsis. PLoS Genet. 2009, 5, e1000492. [Google Scholar] [CrossRef] [PubMed]
- Güsewell, S. Nutrient Resorption of Wetland Graminoids Is Related to the Type of Nutrient Limitation. Funct. Ecol. 2005, 19, 344–354. [Google Scholar] [CrossRef]
- Hu, B.; Jiang, Z.M.; Wang, W.; Qiu, Y.H.; Zhang, Z.H.; Liu, Y.Q.; Li, A.F.; Gao, X.K.; Liu, L.C.; Qian, Y.W.; et al. Nitrate-NRT1.1B-SPX4 Cascade Integrates Nitrogen and Phosphorus Signalling Networks in Plants. Nat. Plants 2019, 5, 401–413. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Yu, Y.M.; Zhang, H.; Long, Y.P.; Shu, Y.; Zhai, J.X. Plant Public RNA-Seq Database: A Comprehensive Online Database for Expression Analysis of ~45 000 Plant Public RNA-Seq Libraries. Plant Biotechnol. J. 2022, 20, 806–808. [Google Scholar] [CrossRef] [PubMed]
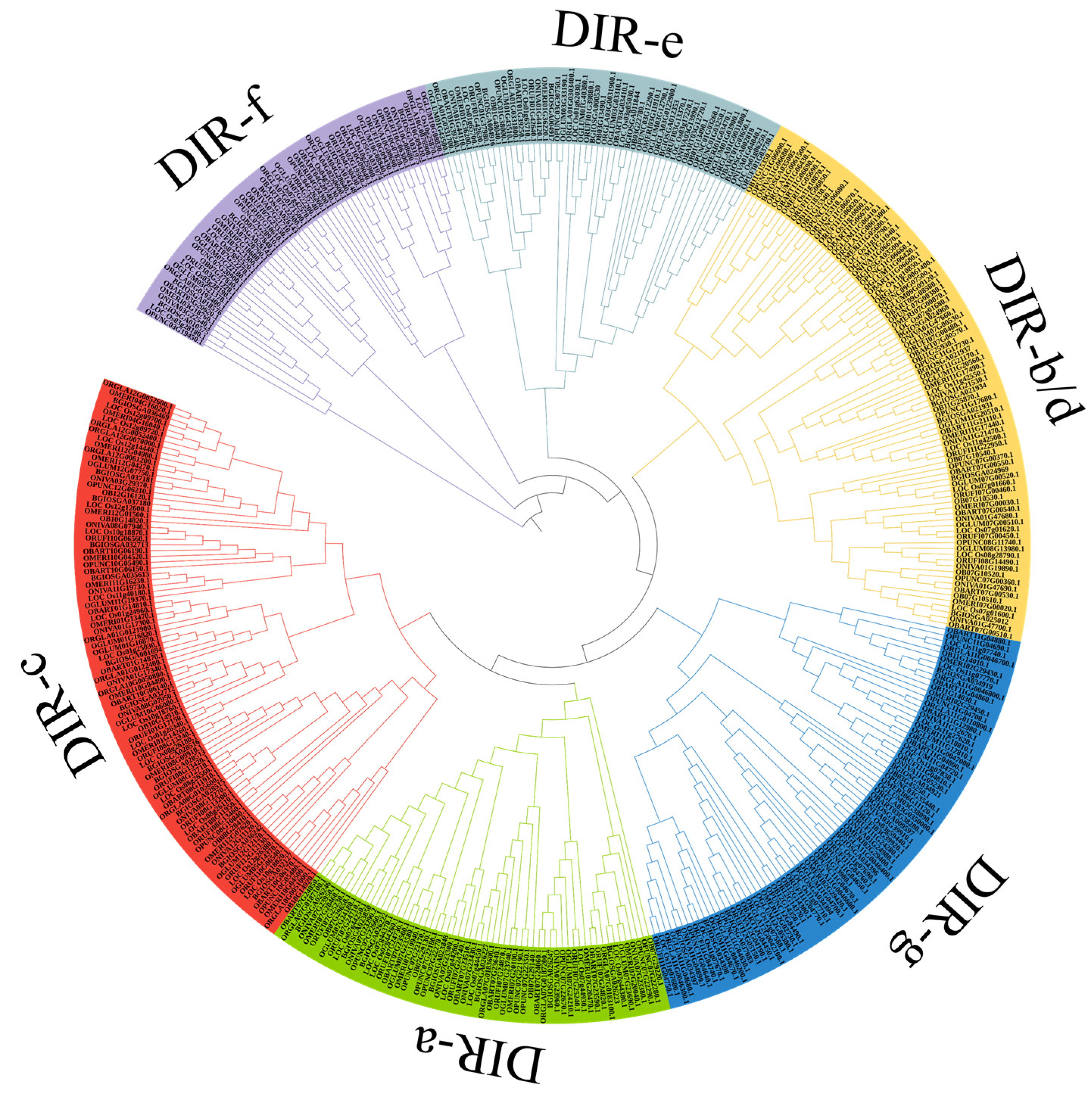
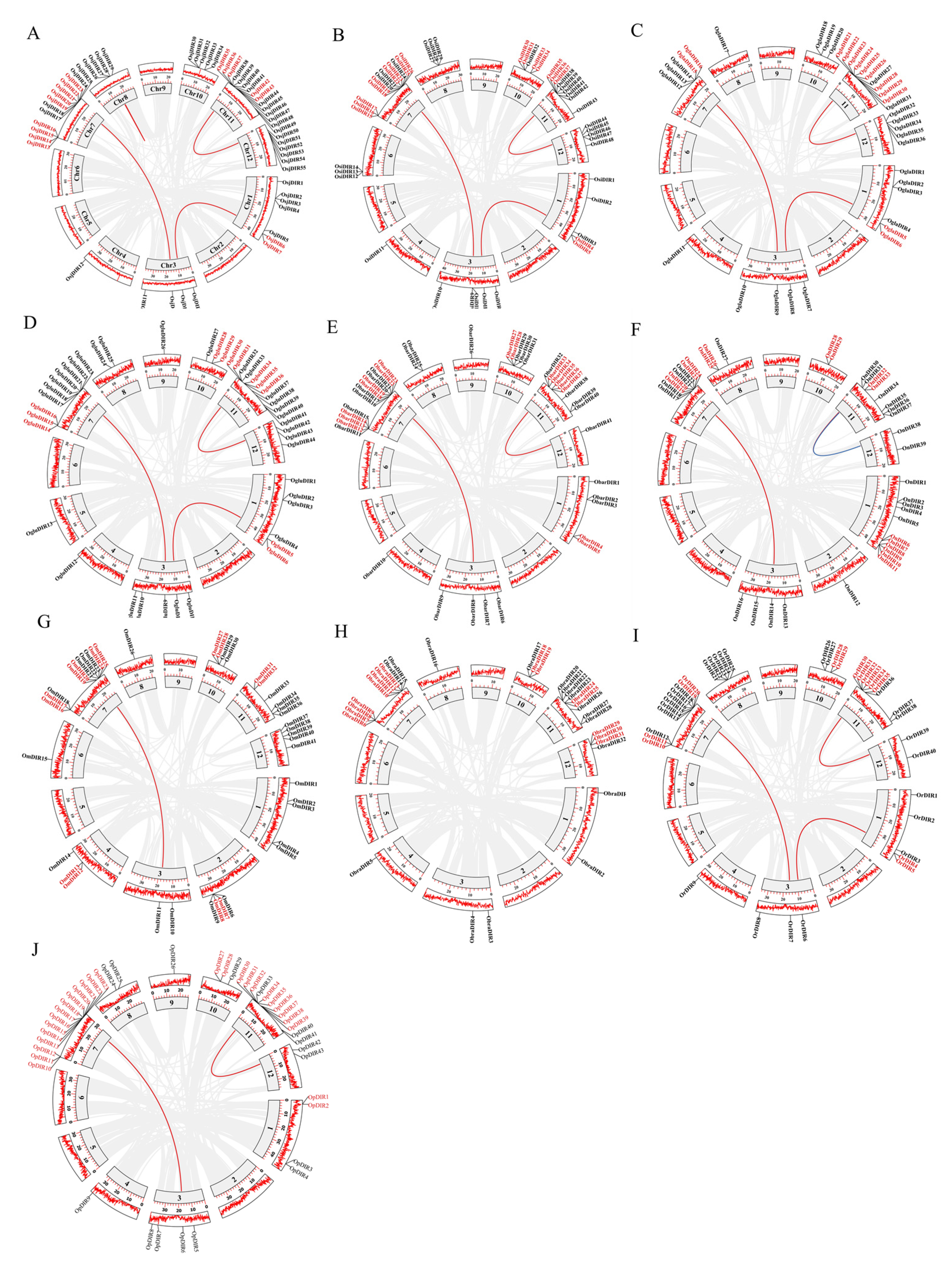
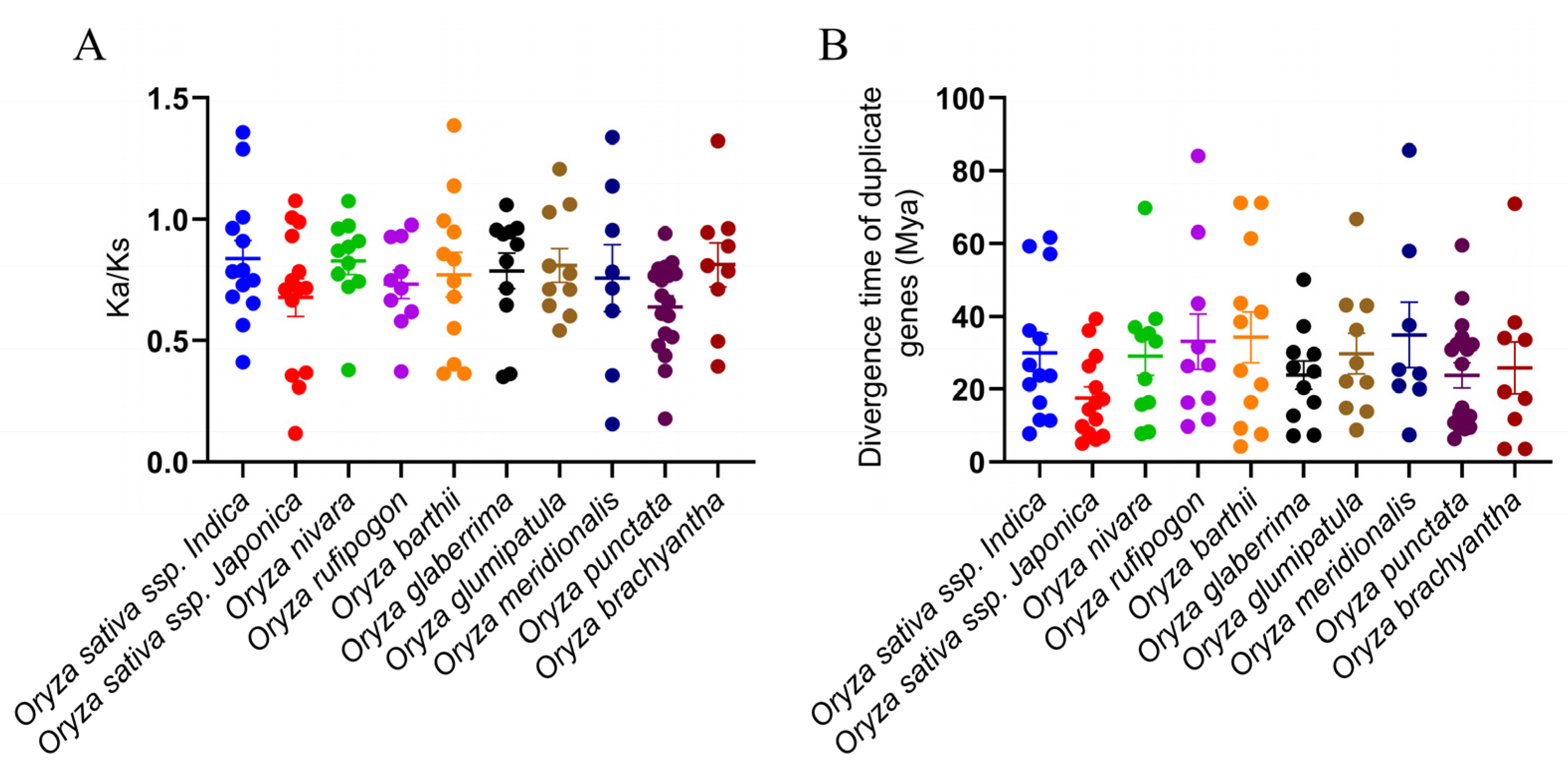

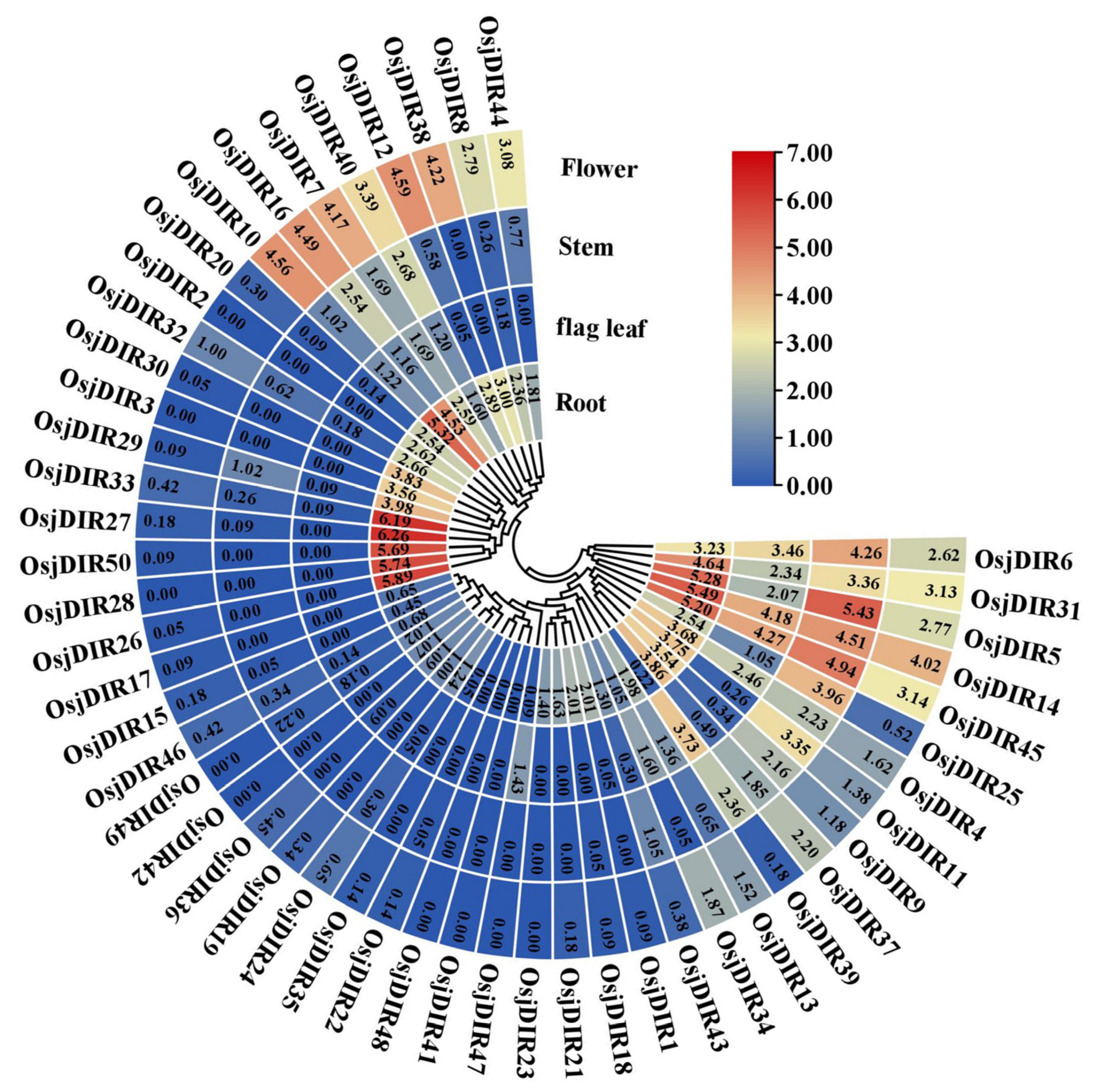
| Species | TD | SD | DIR-a | DIR-c | DIR-e | DIR-f | DIR-g | DIR-b/d | Number |
|---|---|---|---|---|---|---|---|---|---|
| Oryza sativa ssp. indica | 18 | 6 | 7 | 12 | 6 | 7 | 8 | 8 | 48 |
| Oryza sativa ssp. japonica | 17 | 8 | 7 | 15 | 7 | 6 | 10 | 11 | 55 |
| Oryza glaberrima | 12 | 6 | 4 | 9 | 6 | 4 | 10 | 3 | 36 |
| Oryza nivara | 16 | 3 | 6 | 8 | 4 | 6 | 6 | 9 | 39 |
| Oryza rufipogon | 13 | 3 | 7 | 8 | 5 | 6 | 8 | 7 | 40 |
| Oryza barthii | 18 | 4 | 6 | 8 | 5 | 5 | 6 | 11 | 41 |
| Oryza glumipatula | 11 | 6 | 5 | 8 | 7 | 6 | 7 | 11 | 44 |
| Oryza meridionalis | 14 | 2 | 5 | 13 | 4 | 5 | 7 | 7 | 41 |
| Oryza punctata | 26 | 4 | 9 | 4 | 6 | 5 | 8 | 11 | 43 |
| Oryza brachyantha | 15 | 0 | 4 | 5 | 4 | 3 | 7 | 10 | 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, W.; Xue, B.; He, Y.; Liao, S.; Li, X.; Li, X.; Liang, Y.-K. Genome-Wide Identification and Expression Pattern Analysis of Dirigent Members in the Genus Oryza. Int. J. Mol. Sci. 2023, 24, 7189. https://doi.org/10.3390/ijms24087189
Duan W, Xue B, He Y, Liao S, Li X, Li X, Liang Y-K. Genome-Wide Identification and Expression Pattern Analysis of Dirigent Members in the Genus Oryza. International Journal of Molecular Sciences. 2023; 24(8):7189. https://doi.org/10.3390/ijms24087189
Chicago/Turabian StyleDuan, Wen, Baoping Xue, Yaqian He, Shenghao Liao, Xuemei Li, Xueying Li, and Yun-Kuan Liang. 2023. "Genome-Wide Identification and Expression Pattern Analysis of Dirigent Members in the Genus Oryza" International Journal of Molecular Sciences 24, no. 8: 7189. https://doi.org/10.3390/ijms24087189





