Improvement of Salinity Tolerance in Water-Saving and Drought-Resistance Rice (WDR)
Abstract
:1. Introduction
2. Effect of Salt Stress on Rice Growth
3. Salt Tolerance Mechanisms
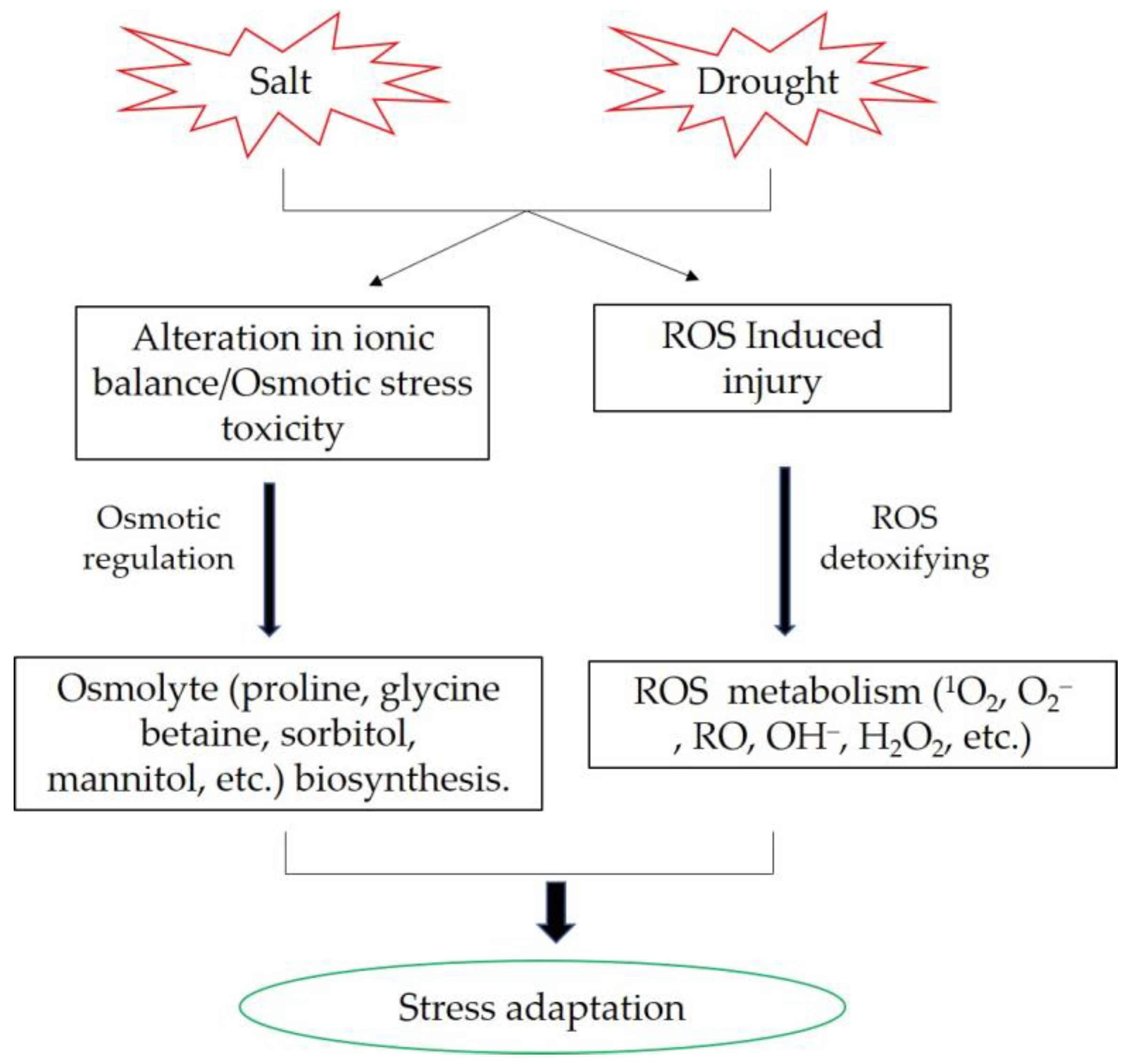
3.1. Osmotic Adjustment
3.2. Ionic Homeostasis Mechanisms
3.3. Resistance to Oxidative Stress
3.4. Signal Molecules
4. Selection and Utilization of Salt-Tolerant Rice Germplasm Resources
5. Strategies for Salt Tolerance Improvement in Rice
5.1. Conventional Breeding
5.2. Molecular Marker-Assisted Selection (MAS)
5.3. Genome Editing
6. Breeding of Salt-Tolerant WDR
- The dominant genic male sterile WDR rice lines can be generated by crossing more than five rounds of backcrossing with the dominant genic male sterile lines (female parents) and the core WDR parents (male parents).
- F1 seeds are obtained by crossing the WDR genic male sterile lines (female parents) with genotypes, such as ‘Haidao 86’, ‘Dongxiang wild rice’, and salt-tolerant donors.
- F1 seeds are mixed and sowed, the rice plants are pollinated during the heading stage, and the seeds are harvested at maturity from the male sterile plants with decent agronomic traits to obtain the first-generation recurrent selection population.
- The first-generation recurrent selection population is planted in a dry land, relying only on rainfall throughout the growth period, without artificial irrigation. At the heading stage, artificial pollination is carried out. The seeds from the elite male sterile plants are harvested at the maturity stage to obtain the generation recurrent selection population.
- The second generation of the recurrent selection population is planted in saline-alkali land or coastal land, and the seeds from the elite male sterile plants are harvested to obtain the third generation of the recurrent selection population.
- According to the breeding objectives and selection pressure, different selection environments can be set to obtain recurrent selection populations, or fertile individual plants that meet the breeding objectives can be selected to conduct conventional breeding programs.
7. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hossain, M.A.; Uddin, M.K.; Ismail, M.R.; Asharafuzzaman, M. Responces of glutamine synthetase-glutamate synthetase cycle enzyme in to tomato under salinity stress. Int. J. Agric. Biol. 2012, 14, 50–515. [Google Scholar]
- FAO Land and Plant Nutrition Management Service. Available online: http://www.fao.org/ag/agl/agll/spush (accessed on 24 May 2010).
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [Green Version]
- Naveed, S.A.; Zhang, F.; Zhang, J.; Zheng, T.Q.; Meng, L.J.; Pang, Y.L.; Xu, J.L.; Li, Z.K. Identification of QTN and candidate genes for salinity tolerance at the germination and seedling stages in rice by genome-wide association analyses. Sci. Rep. 2018, 8, 6505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shahbaz, M.; Ashraf, M. Improving salinity tolerance in cereals. Crit. Rev. Plant Sci. 2013, 32, 237–249. [Google Scholar] [CrossRef]
- Luo, L.; Mei, H.; Yu, X.; Xia, H.; Chen, L.; Liu, H.; Zhang, A.; Xu, K.; Wei, H.; Liu, G.; et al. Water-saving and drought-resistance rice: From the concept to practice and theory. Mol. Breed. 2019, 39, 145. [Google Scholar] [CrossRef]
- Xia, H.; Zhang, X.; Liu, Y.; Bi, J.; Ma, X.; Zhang, A.; Liu, H.; Chen, L.; Zhou, S.; Gao, H.; et al. Blue revolution for food security under carbon neutrality: A case from the water-saving and drought-resistance rice. Mol. Plant 2022, 15, 1401–1404. [Google Scholar] [CrossRef]
- Ponce, K.S.; Meng, L.; Guo, L.; Leng, Y.; Ye, G. Advances in sensing, response and regulation mechanism of salt tolerance in rice. Int. J. Mol. Sci. 2021, 22, 2254. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Cheong, B.E.; Natera, S.; Roessner, U. Morphological and metabolic responses to salt stress of rice (Oryza sativa L.) cultivars which differ in salinity tolerance. Plant Physiol. Biochem. 2019, 144, 427–435. [Google Scholar] [CrossRef]
- Tsai, Y.C.; Chen, K.C.; Cheng, T.S.; Lee, C.; Lin, S.H.; Tung, C.W. Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 2019, 19, 403. [Google Scholar] [CrossRef] [Green Version]
- Xiao, D.; Li, J.; Deng, X.; Wei, P.; Tang, J.; Wei, H.; Chen, Y.; Dai, Q. Response of quality formation of different rice varieties to salt stress. J. Nucl. Agric. Sci. 2020, 34, 1840–1847. [Google Scholar]
- Lin, H.; Zhu, M.; Gao, G.; Liang, Z.; Yano, M.; Su, W.; Hu, X.; Ren, Z.; Chao, D. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor. Appl. Genet. 2004, 108, 253–260. [Google Scholar] [PubMed]
- Garg, A.K. Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl. Acad. Sci. USA 2003, 99, 15898–15903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Sripinyowanich, S.; Klomsaku, L.P.; Boonburapong, B.; Bangyeekhun, T.; Asami, T.; Gu, H.; Buaboocha, T.; Chadchawan, E. Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): The role of Os P5CS1 and Os P5CR gene expression during salt stress. Environ. Exp. Bot. 2013, 86, 94–105. [Google Scholar] [CrossRef]
- Yu, J.; Li, Y.; Tang, W.; Liu, J.; Lu, B.; Liu, Y. The Accumulation of Glycine Betaine Is Dependent on Choline Monooxygenase (OsCMO), Not on Phosphoethanolamine N-Methyltransferase (OsPEAMT1), in Rice (Oryza sativa L. ssp. japonica). Plant. Mol. Biol. Rep. 2014, 32, 916–922. [Google Scholar] [CrossRef]
- Hasthanasombut, S.; Supaibulwatana, K.; Mii, M.; Nakamura, I. Genetic manipulation of Japonica rice using the OsBADH1 gene from Indica rice to improve salinity tolerance. Plant Cell 2011, 104, 79–89. [Google Scholar]
- Li, H.; Zang, B.; Deng, X.; Wang, X. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.; Bohnert, H.J. Plant Cellular and Molecular Responses To High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Chen, S.; Dai, S.; Wang, R.; Li, N.; Shen, X.; Zhou, X.; Lu, C.; Zheng, X.; Hu, Z.; et al. Ion flux profiles and plant ion homeostasis control under salt stress. Plant Signal Behav. 2009, 4, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar]
- Ren, Z.; Gao, J.; Li, L.; Cai, X.; Huang, W.; Chao, D.; Zhu, M.; Wang, Z.; Luan, S.; Lin, H. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.; Zhu, J.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef] [Green Version]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Zheng, L.; Xue, Y.; Zhang, Q.; Wang, L.; Shuo, H. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. J. Plant Biol. 2010, 53, 444–452. [Google Scholar] [CrossRef]
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188. [Google Scholar] [CrossRef]
- Ashraf, M. Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol. Adv. 2009, 27, 84–93. [Google Scholar] [CrossRef]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319. [Google Scholar] [CrossRef]
- Wu, T.; Lin, W.; Kao, C.; Hong, C. Gene knockout of glutathione reductase 3 results in increased sensitivity to salt stress in rice. Plant Mol. Biol. 2015, 87, 555–564. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 2013, 8, e57472. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhou, X.; Tao, M.; Yuan, F.; Liu, L.; Wu, F.; Wu, X.; Xiang, Y.; Niu, Y.; Liu, F.; et al. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 2019, 572, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and finetunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wang, C.; Xue, Y.; Liu, X.; Chen, S.; Song, C.; Yang, Y.; Guo, Y. Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 2019, 10, 1199. [Google Scholar] [CrossRef] [Green Version]
- Lou, L.; Yu, F.; Tian, M.; Liu, G.; Wu, Y.; Wu, Y.; Xia, R.; Pardo, J.; Guo, Y.; Xie, Q. ESCRT-I component VPS23A sustains salt tolerance by strengthening the SOS module in Arabidopsis. Mol. Plant 2020, 13, 1134–1148. [Google Scholar] [CrossRef]
- Li, J.; Zhou, H.; Zhang, Y.; Li, Z.; Yang, Y.; Guo, Y. The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 2020, 55, 367–380. [Google Scholar] [CrossRef]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Song, Y.; Miao, Y.; Song, C. Behind the scenes: The roles of reactive oxygen species in guard cells. New Phytol. 2014, 201, 1121–1140. [Google Scholar] [CrossRef]
- Fernando, L.H. The performance of salt resistant paddy, Pokkali in Ceylon. Trop Agric. 1949, 105, 124–126. [Google Scholar]
- Ghose, R.L.M.; Flowers, W.T. Botanical improvement of varieties-General characters of Indian varieties and the application of genetics to rice improvement. Proc. -Indian Acad. Sci. Sect. B. 1959, 49, 287–302. [Google Scholar] [CrossRef]
- Singh, R.K.; Gregorio, G.B. CSR23: A new salt-tolerant rice variety for India. Int. Rice Res. Notes 2006, 31, 16–18. [Google Scholar] [CrossRef]
- Thomson, M.J.; de Ocampo, M.; Egdane, J.; Rahman, M.A.; Sajise, A.G.; Adorada, D.L.; Tumimbang-Raiz, E.; Blumwald, E.; Seraj, Z.I.; Singh, R.K.; et al. Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice 2010, 3, 148–160. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Yan, G.; Wang, A.; Zhu, G.; Tang, H.; He, C.; Ren, Z.; Liu, K.; Zhang, G.; Shi, W.; et al. Research progress on the breeding of salt-tolerant rice varieties. Barley Cereal Sci. 2017, 34, 1–9. [Google Scholar]
- Hu, S.; Tao, H.; Qian, Q.; Guo, L. Progresses on genetics and molecular breeding for salt-tolerance in rice. Mol. Plant Breed. 2010, 8, 629–640. [Google Scholar]
- Chen, R.; Cheng, Y.; Han, S.; Van Handel, B.; Dong, L.; Li, X.; Xie, X. Whole genome sequencing and comparative transcriptome anal-ysis of a novel seawater adapted, salt-resistant rice cultivar—Sea rice 86. BMC Genom. 2017, 18, 655. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Understanding salinity responses and adopting ‘omics-based’ approaches to generate salinity tolerant cultivars of rice. Front. Plant Sci. 2015, 6, 712. [Google Scholar] [CrossRef] [Green Version]
- Yuan, L. Saline-Alkaline Tolerance Rice Breeding Technology; Shandong Technology Press: Jinan, China, 2019; pp. 121–131. [Google Scholar]
- Das, P.; Mishra, M.; Lakra, N.; Singla-Pareek, S.L.; Pareek, A. Mutation breeding: A powerful approach for obtaining abiotic stress tolerant crops and upgrading food security for human nutrition. In Mutagenesis: Exploring Novel Genes and Pathways; Tomlekova, N.B., Kozgar, M.I., Wani, M.R., Eds.; Wageningen Academic Publisher: Wageningen, The Netherlands, 2014; pp. 615–621. [Google Scholar]
- Mustafa, G.; Soomro, A.M.; Baloch, A.W.; Siddiqui, K.A. “Shua-92,” a new cultivar of rice (Oryza sativa L.) developed through fast neutrons irradiation. Mutat. Breed. Newsl. 1997, 43, 35–36. [Google Scholar]
- Saleem, M.Y.; Mukhtar, Z.; Cheema, A.A.; Atta, B.M. Induced mutation and in vitro techniques as a method to induce salt tolerance in Basmati rice (Oryza sativa L.). Int. J. Environ. Sci. Technol. 2005, 2, 141–145. [Google Scholar] [CrossRef]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Ashraf, M.Y.; Wu, L. Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 1994, 13, 17–42. [Google Scholar] [CrossRef]
- Deng, P.; Jiang, D.; Dong, Y.; Shi, X.; Jing, W.; Zhang, W. Physiological characterization and fine mapping of salt-tolerant mutant in rice (Oryza sativa). Funct. Plant Biol. 2015, 42, 1026–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganapati, R.K.; Naveed, S.A.; Zafar, S.; Wang, W.; Xu, J. Saline-Alkali Tolerance in Rice: Physiological Response, Molecular Mechanism, and QTL Identification and Application to Breeding. Rice Sci. 2022, 29, 412–434. [Google Scholar] [CrossRef]
- Singh, R.; Singh, Y.; Xalaxo, S.; Verulkar, S.; Yadav, N.; Singh, S.; Singh, N.; Prasad, K.S.N.; Kondayya, K.; Rao, P.V.R.; et al. From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci. 2016, 242, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Vinod, K.K.; Krishnan, S.G.; Babu, N.N.; Nagarajan, M.; Singh, A.K. Improving salt tolerance in rice: Looking beyond the conventional. In Salt Stress in Plants: Signalling, Omics and Adaptations; Ahmad, P., Azooz, M.M., Prasas, M.N., Eds.; Springer: New York, NY, USA, 2013; pp. 219–260. [Google Scholar]
- Fan, X.; Jiang, H.; Meng, L.; Chen, J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 11674. [Google Scholar] [CrossRef]
- Gregorio, G.B.; Senadhira, D. Genetic analysis of salinity tolerance in rice. Theor. Appl. Genet. 1993, 86, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Li, Y.; Huang, R. Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef]
- Huyen, L.T.N.; Cuc, L.M.; Ismail, A.M.; Ham, L.H. Introgression the Salinity Tolerance QTLs Saltol into AS996, the Elite Rice Variety of Vietnam. Am. J. Plant Sci. 2012, 3, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Bimpong, I.K.; Manneh, B.; Sock, M.; Diaw, F.; Amoah, N.K.A.; Ismail, A.M.; Gregorio, G.; Singh, R.K.; Wopereis, M. Improving salt tolerance of lowland rice cultivar ‘Rassi’ through marker-aided backcross breeding in West Africa. Plant Sci. 2016, 242, 288–299. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Gopalakrishnan, S.; Singh, V.P.; Prabhu, K.V.; Mohapatra, T.; Singh, N.K.; Sharma, T.R.; Nagarajan, M.; Ellur, R.K.; Singh, A.; et al. Marker assisted selection: A paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 2011, 71, 120. [Google Scholar]
- Geetha, S.; Vasuki, A.; Selvam, P.J.; Saraswathi, R.; Krishnamurthy, S.L.; Dhasarathan, M.; Thamodharan, G.; Baskar, M. Development of sodicity tolerant rice varieties through marker assisted backcross breeding. Electron. J. Plant Breed. 2017, 8, 1013. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Okporie, E.; Onyishi, G.; Utobo, E.; Ekwu, L.; Swaray, S.; et al. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 2019, 33, 440–455. [Google Scholar] [CrossRef] [Green Version]
- Shailani, A.; Joshi, R.; Singla-Pareek, S.L.; Pareek, A. Stacking for future: Pyramiding genes to improve drought and salinity tolerance in rice. Physiol. Plant. 2021, 172, 1352–1362. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, J.; Yu, D.; Xu, P. Identification and validation a major QTL from“Sea Rice 86” seedlings conferred salt tolerance. Agronomy 2020, 10, 410. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of Targeted Point Mutations in Rice by a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef] [Green Version]
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome Editing in Rice: Recent Advances, Challenges, and Future Implications. Front. Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chao, D.; Gao, J.; Zhu, M.; Shi, M.; Lin, H. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef] [Green Version]
- Santosh-Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.K.; Xu, J.L. Breeding for Drought and Salt Tolerant Rice (Oryza Sativa L.): Progress and Perspectives. In Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 531–564. [Google Scholar]
- Luo, L. Breeding for water-saving and drought-resistance rice (WDR) in China. J. Exp. Bot. 2010, 61, 3509–3517. [Google Scholar] [CrossRef] [Green Version]
- Blum, A. Drought resistance, water-use efficiency, and yield potential–are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 2005, 25, 1159–1168. [Google Scholar] [CrossRef] [Green Version]
- Pennisi, E. The blue revolution, drop by drop, gene by gene. Science 2008, 320, 171–173. [Google Scholar] [CrossRef]
- Wei, H.; Feng, F.; Lou, Q.; Xia, H.; Ma, X.; Liu, Y.; Xu, K.; Yu, X.; Mei, H.; Luo, L. Genetic determination of the enhanced drought resistance of rice maintainer HuHan2B by pedigree breeding. Sci. Rep. 2016, 6, 37302. [Google Scholar] [CrossRef] [Green Version]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Akram, N.A. Improving salinity tolerance of plants through conventional breeding and genetic engineering: An analytical comparison. Biotechnol. Adv. 2009, 27, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Sahoo, K.; Singh, A.; Anwar, K.; Pundir, P.; Gautam, R.; Krishnamurthy, S.; Sopory, S.; Pareek, A.; Singla-Pareek, S. Enhancing trehalose biosynthesis improves yield potential in marker-free transgenic rice under drought, saline, and sodic conditions. J. Exp. Bot. 2020, 71, 653–668. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.; Liu, A.; Zou, D.; Chen, X. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, N.; Ma, X.; Zhou, H.; Cui, Y.; Wang, C.; Xu, G. Overexpression of OsRLCK241 confers enhanced salt and drought tolerance in transgenic rice (Oryza sativa L.). Gene 2021, 768, 145278. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, F. Rice breeding in the post-genomics era: From concept to practice. Curr. Opin. Plant Biol. 2013, 16, 261–269. [Google Scholar] [CrossRef]
- Ali, A.; Xu, J.; Ismail, A.; Fu, B.; Vijaykumar, C.; Gao, Y.; Domingo, J.; Maghirang, R.; Yu, S.; Gregorio, G.; et al. Hidden diversity for abiotic and biotic stress tolerances in the primary gene pool of rice revealed by a large backcross breeding program. Field Crops Res. 2006, 97, 66–76. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, F.; Luo, X.; Liu, Y.; Zhang, F.; Liu, G.; Yu, X.; Luo, L. Germplasm enhancement of water-saving and drought-resistance rice based on recurrent selection facilitated by dominant nucleus male sterility. Acta Agric. 2022, 38, 91–95. [Google Scholar]
- Liu, G.; Chen, Z.; Zhang, Z.; Wang, J.; Yu, X.; Luo, L. Germplasm innovation of water-saving and drought-resistance rice with saline-alkaline tolerance. Acta Agric. 2022, 38, 96–102. [Google Scholar]

| Lines | Plant Height (cm) | No. of Effective Spikes per Plant | Spike Length (cm) | Seeding Rate (%) | 1000 Grain Weight (g) | Yield per Plant (g) | Salt Stress Index (%) a | Salt Tolerance Score b | Evaluation of Salt Tolerance c | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Salt Treatment | Normal Control | Salt Treatment | Normal Control | Salt Treatment | Normal Control | Salt Treatment | Normal Control | Salt Treatment | Normal Control | Salt Treatment | Normal Control | ||||
| NYS5 | 74.0 | 81.0 | 12.0 | 14.3 | 17.3 | 18.7 | 68.1 | 82.6 | 20.8 | 22.7 | 12.9 | 15.6 | 17.2 | 1 | HR |
| NYS6 | 93.0 | 104.6 | 14.6 | 18.6 | 15.7 | 17.3 | 60.1 | 80.7 | 21.5 | 24.7 | 8.6 | 12.3 | 30.1 | 2 | R |
| NYS8 | 99.1 | 122.4 | 6.2 | 11.2 | 20.9 | 21.4 | 76.0 | 92.1 | 18.7 | 23.2 | 9.6 | 14.9 | 35.2 | 2 | R |
| NYS10 | 83.2 | 96.8 | 10.0 | 13.8 | 17.9 | 19.1 | 63.6 | 85.0 | 18.9 | 23.0 | 6.7 | 8.6 | 22.1 | 2 | R |
| NYS11 | 74.6 | 81.5 | 7.4 | 10.4 | 16.6 | 18.4 | 45.1 | 70.8 | 15.7 | 21.4 | 7.4 | 10.3 | 28.3 | 2 | R |
| NYS15 | 79.1 | 86.3 | 4.4 | 7.0 | 19.0 | 19.8 | 63.8 | 79.4 | 18.6 | 20.5 | 13.5 | 18.5 | 26.9 | 2 | R |
| NYS16 | 65.8 | 68.2 | 8.6 | 12.0 | 11.7 | 12.1 | 49.6 | 76.2 | 18.7 | 25.9 | 8.0 | 14.0 | 42.9 | 3 | MR |
| NYS17 | 69.6 | 76.2 | 6.2 | 8.2 | 18.1 | 19.1 | 78.1 | 91.5 | 17.5 | 21.3 | 9.0 | 11.5 | 21.5 | 2 | R |
| NYS23 | 67.1 | 78.3 | 10.0 | 14.8 | 12.3 | 13.7 | 58.5 | 77.4 | 16.3 | 22.1 | 8.3 | 20.3 | 59.1 | 3 | MR |
| NYS39 | 84.8 | 102.7 | 7.2 | 8.6 | 19.1 | 21.0 | 82.5 | 95.8 | 18.0 | 19.6 | 12.0 | 17.5 | 31.3 | 2 | R |
| NYS55 | 71.2 | 88.5 | 4.0 | 5.6 | 19.3 | 20.2 | 74.9 | 83.8 | 17.4 | 19.8 | 10.4 | 16.7 | 37.7 | 2 | R |
| NYS73 | 77.7 | 95.1 | 7.8 | 9.8 | 19.2 | 20.6 | 73.7 | 79.1 | 18.2 | 22.7 | 11.2 | 25.5 | 56.2 | 3 | MR |
| NYS83 | 72.4 | 84.7 | 10.8 | 13.8 | 16.7 | 18.1 | 65.6 | 83.6 | 19.6 | 22.8 | 18.6 | 23.3 | 20.4 | 2 | R |
| NYS87 | 75.0 | 88.5 | 8.0 | 13.0 | 21.1 | 19.4 | 60.8 | 93.7 | 20.0 | 21.2 | 8.6 | 14.3 | 39.5 | 2 | R |
| NYS92 | 81.0 | 99.3 | 8.0 | 10.8 | 20.4 | 21.4 | 80.6 | 93.7 | 19.0 | 21.6 | 12.6 | 16.7 | 24.7 | 2 | R |
| NYS101 | 92.4 | 111.8 | 5.6 | 7.2 | 19.5 | 20.8 | 67.0 | 92.0 | 17.4 | 18.9 | 11.6 | 19.7 | 41.0 | 3 | MR |
| NYS105 | 79.9 | 99.5 | 7.0 | 10.8 | 18.1 | 20.2 | 73.7 | 87.5 | 16.3 | 18.9 | 10.2 | 14.1 | 27.7 | 2 | R |
| NYS117 | 67.6 | 79.6 | 7.2 | 9.2 | 18.0 | 19.4 | 71.5 | 87.0 | 17.1 | 20.8 | 8.5 | 10.3 | 18.2 | 1 | HR |
| NYS120 | 77.6 | 108.7 | 8.8 | 12.2 | 18.4 | 21.5 | 68.9 | 84.3 | 19.1 | 22.1 | 7.2 | 14.6 | 50.4 | 3 | MR |
| Huhan 61 | 62.3 | 68.6 | 9.3 | 9.4 | 12.4 | 12.7 | 42.9 | 79.8 | 18.2 | 25.2 | 6.8 | 16.2 | 58.2 | 3 | MR |
| Huhan 3 | 60.3 | 73.3 | 6.8 | 9.4 | 12.2 | 12.6 | 48.5 | 81.0 | 18.7 | 25.2 | 7.8 | 13.6 | 42.6 | 3 | MR |
| Sea Rice 86 | 93.0 | 105.7 | 16.0 | 20.3 | 15.3 | 17.1 | 65.7 | 77.4 | 22.1 | 24.8 | 12.7 | 15.6 | 18.8 | 1 | HR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, F.; Zhang, A.; Chen, Z.; Luo, X.; Kong, D.; Zhang, F.; Yu, X.; Liu, G.; Luo, L. Improvement of Salinity Tolerance in Water-Saving and Drought-Resistance Rice (WDR). Int. J. Mol. Sci. 2023, 24, 5444. https://doi.org/10.3390/ijms24065444
Liu Y, Wang F, Zhang A, Chen Z, Luo X, Kong D, Zhang F, Yu X, Liu G, Luo L. Improvement of Salinity Tolerance in Water-Saving and Drought-Resistance Rice (WDR). International Journal of Molecular Sciences. 2023; 24(6):5444. https://doi.org/10.3390/ijms24065444
Chicago/Turabian StyleLiu, Yi, Feiming Wang, Anning Zhang, Zhihao Chen, Xingxing Luo, Deyan Kong, Fenyun Zhang, Xinqiao Yu, Guolan Liu, and Lijun Luo. 2023. "Improvement of Salinity Tolerance in Water-Saving and Drought-Resistance Rice (WDR)" International Journal of Molecular Sciences 24, no. 6: 5444. https://doi.org/10.3390/ijms24065444






