Domain Swapping between AtACS7 and PpACL1 Results in Chimeric ACS-like Proteins with ACS or Cβ-S Lyase Single Enzymatic Activity
Abstract
:1. Introduction
2. Results
2.1. Replacing Box 6 of AtACS7 with That of PpACL1 Led to a Loss of ACS Activity but Retains CSL Activity In Vitro
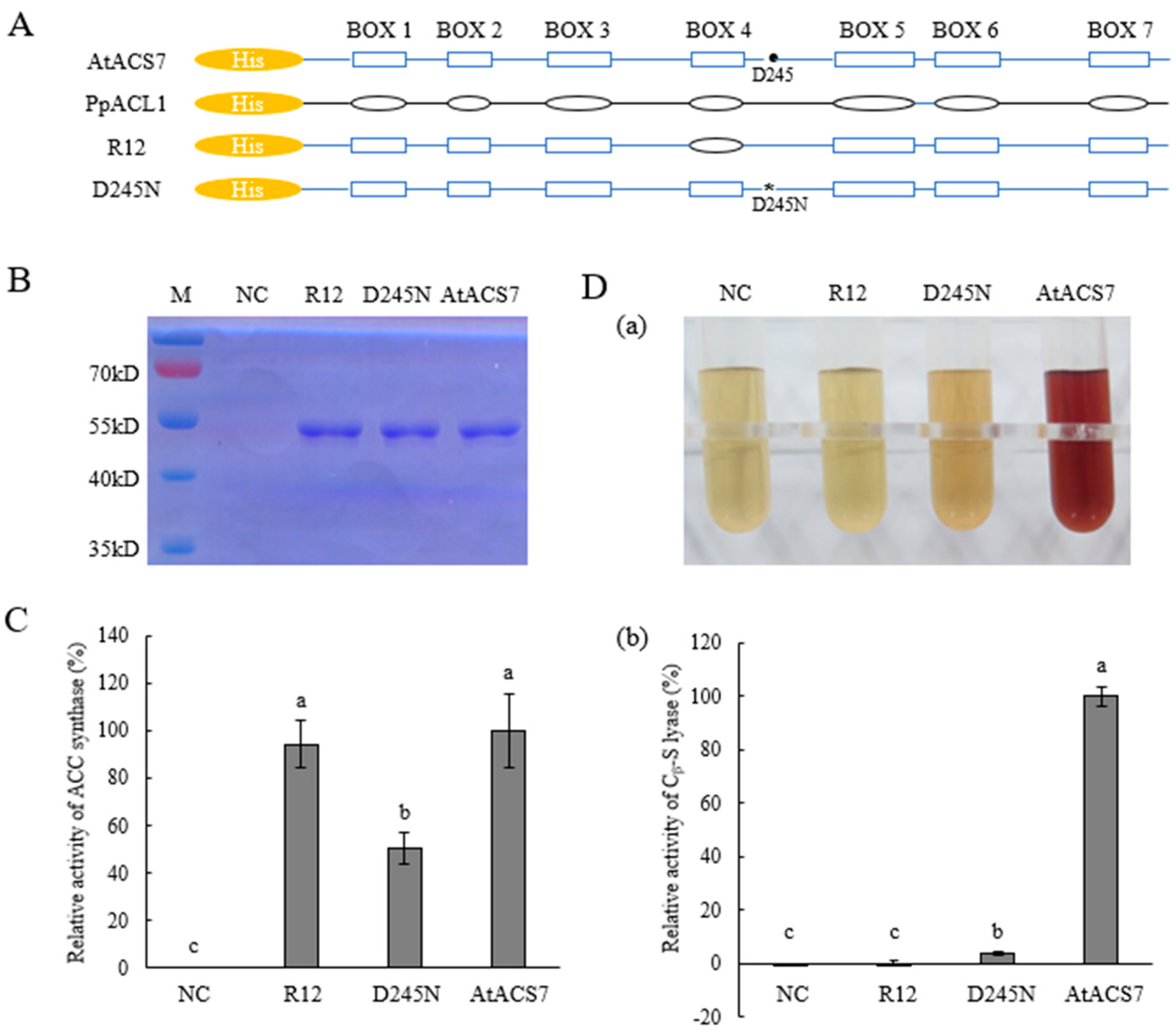
2.2. Substituting Box 4 of AtACS7 with That of PpACL1 Resulted in the Chimeric R12 Protein Having Only ACS Single Activity
3. Discussion
4. Materials and Methods
4.1. Construction and Purification of Chimeric Proteins R10 and R12
4.2. In Vitro ACS Activity Measurement
4.3. In Vitro Cβ-S Lyase Activity
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Van de Poel, B.; Cooper, E.D.; Thierer, J.H.; Gibbons, T.R.; Delwiche, C.F.; Chang, C. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 2015, 1, 14004. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Van den Broeck, L.; Inze, D. The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Cellini, A.; Donati, I.; Farneti, B.; Khomenko, I.; Buriani, G.; Biasioli, F.; Cristescu, S.M.; Spinelli, F. A Breach in Plant Defences: Pseudomonas syringae pv. actinidiae Targets Ethylene Signalling to Overcome Actinidia chinensis Pathogen Responses. Int. J. Mol. Sci. 2021, 22, 4375. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Trivellini, A.; Chhillar, H.; Chopra, P.; Ferrante, A.; Khan, N.A.; Ismail, A.M. The significance and functions of ethylene in flooding stress tolerance in plants. Environ. Exp. Bot. 2020, 179, 104188. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Burger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Yang, S.F.; Hoffman, N.E. Ethylene Biosynthesis and its Regulation in Higher Plants. Ann. Rev. Plant Physiol. 1984, 35, 155–189. [Google Scholar] [CrossRef]
- Alexander, F.W.; Sandmeier, E.; Mehta, P.K.; Christen, P. Evolutionary relationships among pyridoxal-5’-phosphate-dependent enzymes. Regio-specific alpha, beta and gamma families. Eur. J. Biochem. 1994, 219, 953–960. [Google Scholar] [CrossRef]
- Mehta, P.K.; Christen, P. Homology of 1-aminocyclopropane-1-carboxylate synthase, 8-amino-7-oxononanoate synthase, 2-amino-6-caprolactam racemase, 2,2-dialkylglycine decarboxylase, glutamate-1-semialdehyde 2,1-aminomutase and isopenicillin-N-epimerase with aminotransferases. Biochem. Biophys. Res. Commun. 1994, 198, 138–143. [Google Scholar] [CrossRef]
- Mehta, P.K.; Christen, P. The molecular evolution of pyridoxal-5’-phosphate-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 2000, 74, 129–184. [Google Scholar] [PubMed]
- Zhang, T.C.; Qiao, Q.; Zhong, Y. Detecting adaptive evolution and functional divergence in aminocyclopropane-1-carboxylate synthase (ACS) gene family. Comput. Biol. Chem. 2012, 38, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Geck, M.K.; Eliot, A.C.; Kirsch, J.F. Aminotransferase activity and bioinformatic analysis of 1-aminocyclopropane-1-carboxylate synthase. Biochemistry 2000, 39, 15242–15249. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Dong, H.; Nasrullah; Mei, Y.; Wang, N.N. Functional investigation of two 1-aminocyclopropane-1-carboxylate (ACC) synthase-like genes in the moss Physcomitrella patens. Plant Cell Rep. 2016, 35, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Hao, B.; Sun, G.; Mei, Y.; Sun, L.; Sun, Y.; Wang, Y.; Zhang, Y.; Zhang, W.; Zhang, M.; et al. Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes. Sci. Adv. 2021, 7, eabg8752. [Google Scholar] [CrossRef]
- Yang, S. Faculty Opinions Recommendation of [Xu C et al., Sci Adv 2022 7(46:eabg8752)]. Faculty Opinions. 2022. Available online: https://facultyopinions.com/article/741145724 (accessed on 15 May 2022).
- Dong, J.G.; Kim, W.T.; Yip, W.K.; Thompson, G.A.; Li, L.; Bennett, A.B.; Yang, S.F. Cloning of a cDNA encoding 1-aminocyclopropane-1-carboxylate synthase and expression of its mRNA in ripening apple fruit. Planta 1991, 185, 38–45. [Google Scholar] [CrossRef]
- Yip, W.K.; Dong, J.G.; Kenny, J.W.; Thompson, G.A.; Yang, S.F. Characterization and sequencing of the active site of 1-aminocycloproane-1-carboxylate synthase. Proc. Natl. Acad. Sci. USA 1990, 87, 7930–7934. [Google Scholar] [CrossRef]
- Tsuchisaka, A.; Yu, G.; Jin, H.; Alonso, J.M.; Ecker, J.R.; Zhang, X.; Gao, S.; Theologis, A. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana. Genetics 2009, 183, 979–1003. [Google Scholar] [CrossRef]
- Huai, Q.; Xia, Y.; Chen, Y.; Callahan, B.; Li, N.; Ke, H. Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5’-phosphate provide new insight into catalytic mechanisms. J. Biol. Chem. 2001, 276, 38210–38216. [Google Scholar] [CrossRef]
- Li, J.F.; Qu, L.H.; Li, N. Try152 plays central role in the catalysis of 1-aminocyclopropane-1-carboxylate synthase. J. Exp. Bot. 2005, 56, 2203–2210. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.L.; Capitani, G.; Feng, L.; Gruetter, M.G.; Kirsch, J.F. Glutamate 47 in 1-aminocyclopropane-1-carboxylate synthase is a major specificity determinant. Biochemistry 2001, 40, 12276–12284. [Google Scholar] [PubMed]
- Yamagami, T.; Tsuchisaka, A.; Yamada, K.; Haddon, W.F.; Harden, L.A.; Theologis, A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 2003, 278, 49102–49112. [Google Scholar] [PubMed]
- Le, X.; Lee, C.P.; Monachello, D.; Millar, A.H. Metabolic evidence for distinct pyruvate pools inside plant mitochondria. Nat. Plants 2022, 8, 694–705. [Google Scholar] [PubMed]
- Zhang, Y.; Taufalele, P.V.; Cochran, J.D.; Robillard-Frayne, I.; Marx, J.M.; Soto, J.; Rauckhorst, A.J.; Tayyari, F.; Pewa, A.D.; Gray, L.R.; et al. Mitochondrial pyruvate carriers are required for myocardial stress adaptation. Nat. Metab. 2020, 2, 1248–1264. [Google Scholar] [PubMed]
- Aiyar, A.; Xiang, Y.; Leis, J. Site-Directed Mutagenesis Using Overlap Extension PCR. In In Vitro Mutagenesis Protocols; Trower, M.K., Ed.; Humana Press: Totowa, NJ, USA, 1996; Volume 57, pp. 177–191. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Sun, L.; Mei, Y.; Sun, G.; Li, W.; Wang, D.; Li, X.; Wang, N.N. Domain Swapping between AtACS7 and PpACL1 Results in Chimeric ACS-like Proteins with ACS or Cβ-S Lyase Single Enzymatic Activity. Int. J. Mol. Sci. 2023, 24, 2956. https://doi.org/10.3390/ijms24032956
Xu C, Sun L, Mei Y, Sun G, Li W, Wang D, Li X, Wang NN. Domain Swapping between AtACS7 and PpACL1 Results in Chimeric ACS-like Proteins with ACS or Cβ-S Lyase Single Enzymatic Activity. International Journal of Molecular Sciences. 2023; 24(3):2956. https://doi.org/10.3390/ijms24032956
Chicago/Turabian StyleXu, Chang, Lifang Sun, Yuanyuan Mei, Gongling Sun, Wenjing Li, Dan Wang, Xin Li, and Ning Ning Wang. 2023. "Domain Swapping between AtACS7 and PpACL1 Results in Chimeric ACS-like Proteins with ACS or Cβ-S Lyase Single Enzymatic Activity" International Journal of Molecular Sciences 24, no. 3: 2956. https://doi.org/10.3390/ijms24032956




