Characterization of the IAA-Producing and -Degrading Pseudomonas Strains Regulating Growth of the Common Duckweed (Lemna minor L.)
Abstract
:1. Introduction
2. Results
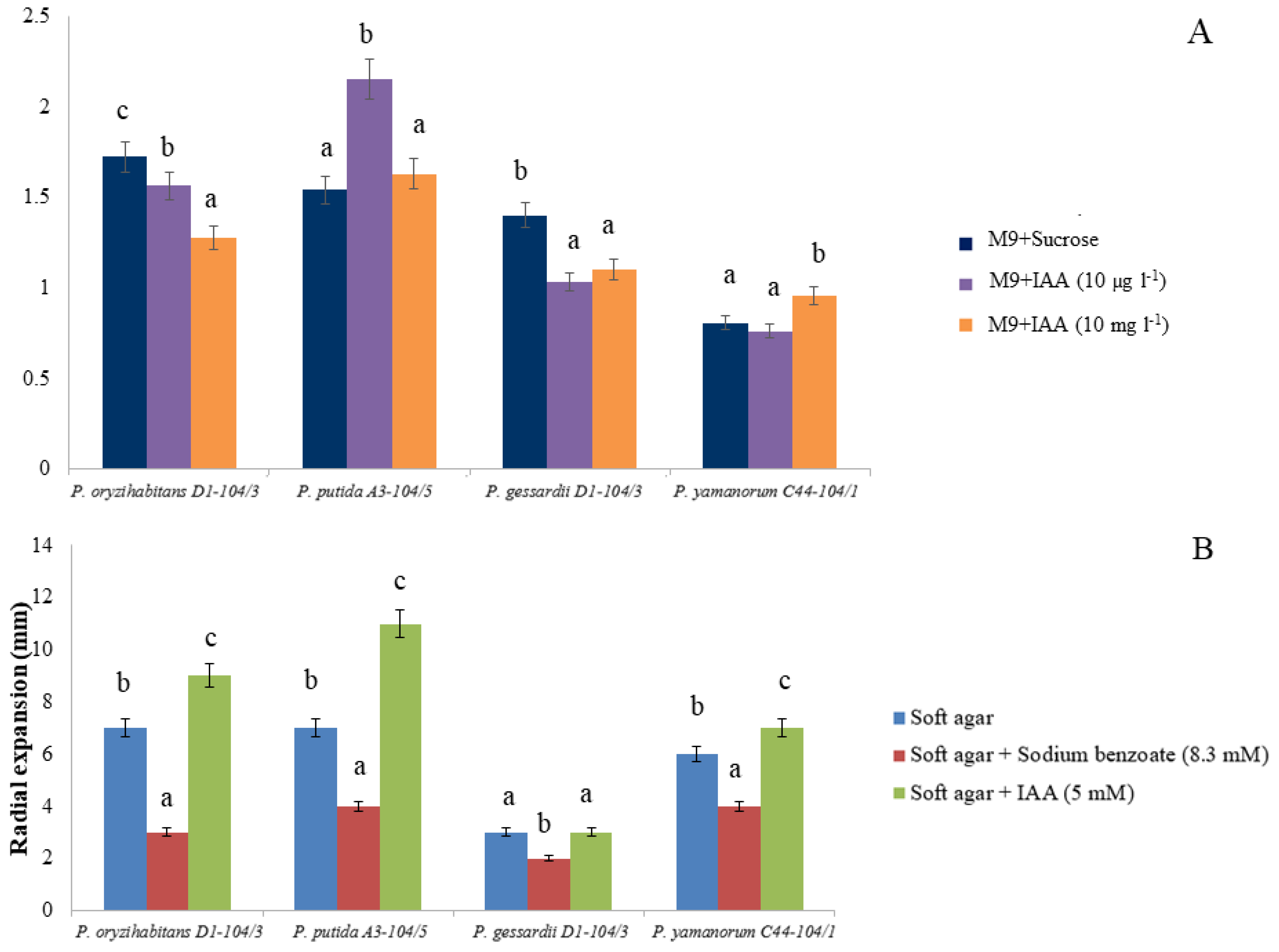
2.1. Analysis of Bacterial Ability to Utilize IAA as A Sole Carbon Source and Evolutionary Relations between IAA-Producing and -Utilizing Bacterial Strains
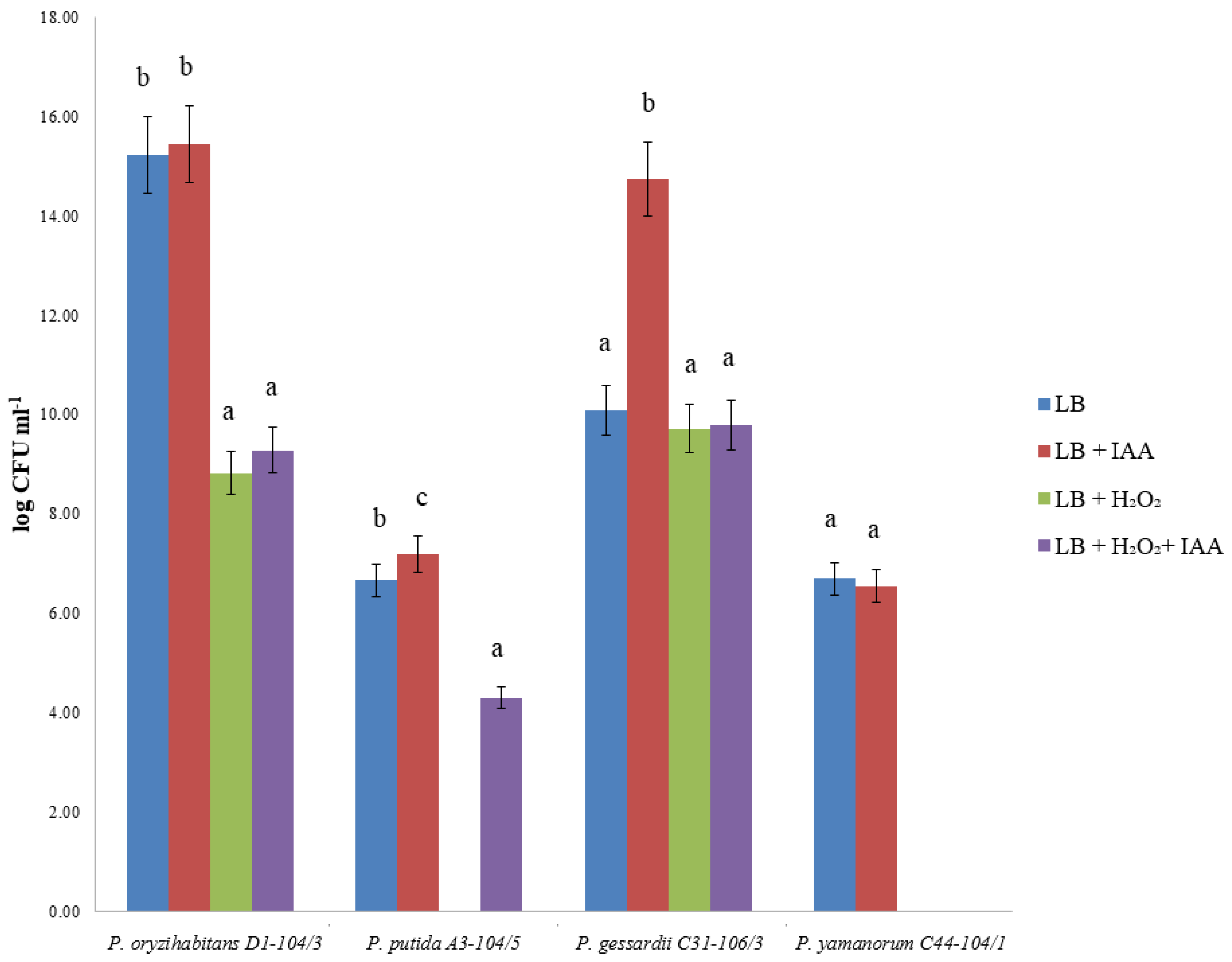
2.2. Bacterial Viability after H2O2 and IAA Treatment
2.3. Biofilm Formation and Chemotaxis
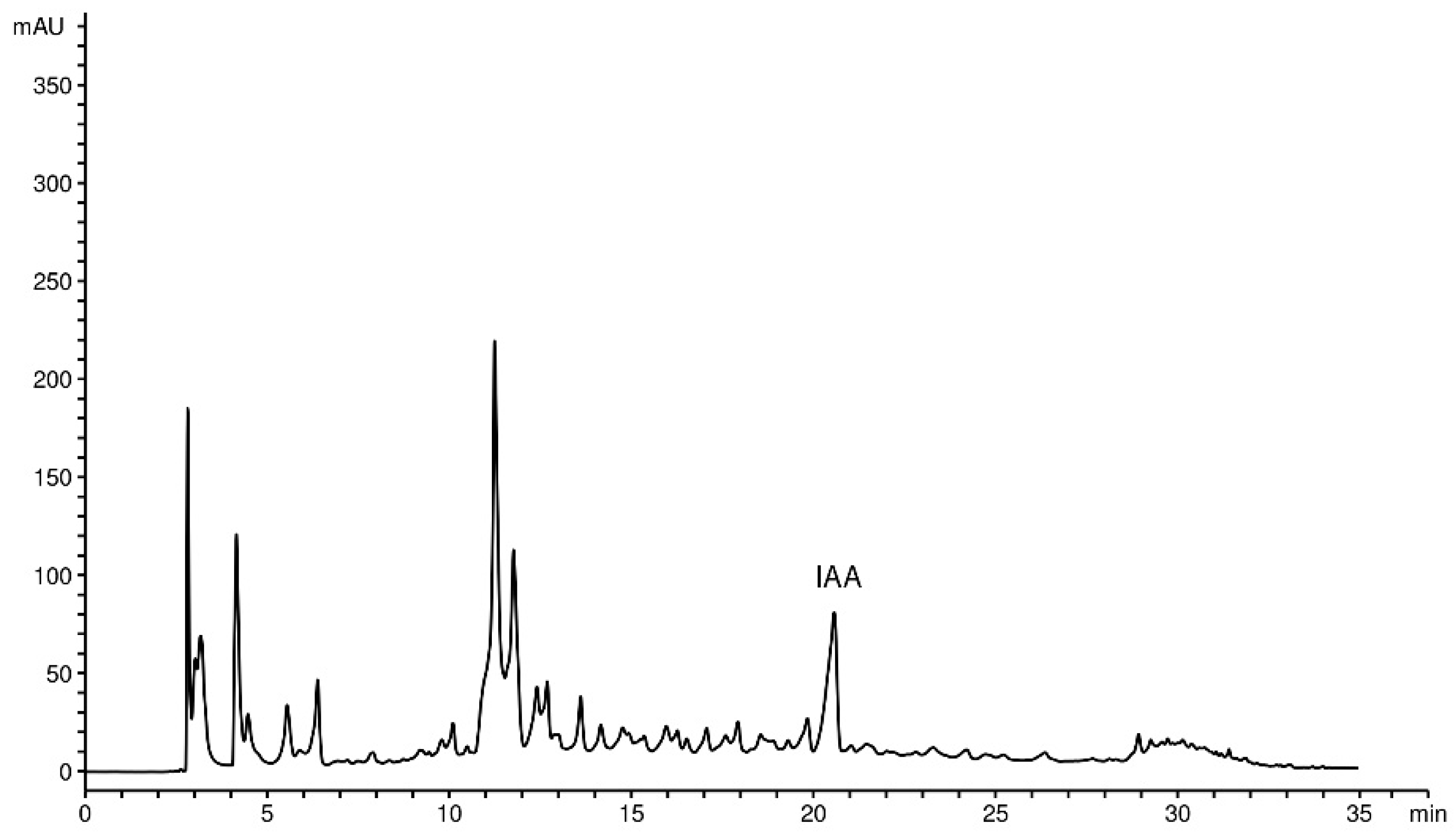
2.4. HPLC-DAD Determination of IAA
2.5. Co-Cultivation of Bacteria and Duckweeds
3. Discussion
3.1. Bacterial Production of IAA
3.2. Bacterial Degradation of IAA
3.3. IAA and Bacterial Viability under Oxidative Stress, Biofilm Formation and Chemotaxis
3.4. Effects of Bacterial Strains and Exogenous IAA on Doubling Time of Duckweeds
4. Material and Methods
4.1. Plant Material and Growth Conditions
4.2. Initial IAA Screening of Bacterial Isolates Associated with the Rhizosphere of L. minor: The Microwell Method
4.3. Bacterial Growth in Minimal M9 Medium with IAA as the Sole Carbon Source
4.4. Bacterial Degradation of IAA
4.5. Bacterial Viability after H2O2 and IAA Treatment
4.6. Biofilm Formation
4.7. Soft Agar Swim Assay
4.8. Extract Preparation and HPLC-DAD Determination of IAA
4.9. Co-Cultivation of Selected Bacterial Strains and Duckweeds
4.10. Phylogenetic Analysis
4.11. Statistical Analysis and Graphical Presentation of the Results
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant–microbe interactions. Antonie Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Xu, J.; Acosta, K.; Poulev, A.; Lebeis, S.; Lam, E. Bacterial Production of Indole Related Compounds Reveals Their Role in Association Between Duckweeds and Endophytes. Front. Chem. 2018, 6, 265. [Google Scholar] [CrossRef] [PubMed]
- Makino, A.; Nakai, R.; Yoneda, Y.; Toyama, T.; Tanaka, Y.; Meng, X.-Y.; Mori, K.; Ike, M.; Morikawa, M.; Kamagata, Y.; et al. Isolation of Aquatic Plant Growth-Promoting Bacteria for the Floating Plant Duckweed (Lemna minor). Microorganisms 2022, 10, 1564. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J. Auxin and Plant-Microbe Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Srivastava, J.K.; Chandra, H.; Kalra, S.J.S.; Mishra, P.; Khan, H.; Yadav, P. Plant–microbe interaction in aquatic system and their role in the management of water quality: A review. Appl. Water Sci. 2016, 7, 1079–1090. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R.; Peters, E.G.; Fei, H.; Papadopoulos, Y.A.; Vessey, J.K.; Ahmad, M.; Asghar, H.N.; Asghar, M.; Gamalero, E.; et al. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996, 42, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Leveau, J.H.J.; Lindow, S.E. Utilization of the Plant Hormone Indole-3-Acetic Acid for Growth by Pseudomonas putida Strain 1290. Appl. Environ. Microbiol. 2005, 71, 2365–2371. [Google Scholar] [CrossRef]
- Laird, T.S.; Flores, N.; Leveau, J.H.J. Bacterial catabolism of indole-3-acetic acid. Appl. Microbiol. Biotechnol. 2020, 104, 9535–9550. [Google Scholar] [CrossRef]
- Ware, A.; Jones, D.H.; Flis, P.; Chrysanthou, E.; Smith, K.E.; Kümpers, B.M.; Yant, L.; Atkinson, J.A.; Wells, D.M.; Bhosale, R.; et al. Loss of ancestral function in duckweed roots is accompanied by progressive anatomical reduction and a re-distribution of nutrient transporters. Curr. Biol. 2023, 33, 1795–1802.e4. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Lee, J.; Lee, J. Diverse roles of microbial indole compounds in eukaryotic systems. Biol. Rev. 2021, 96, 2522–2545. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, W. Indole: A signaling molecule or a mere metabolic byproduct that alters bacterial physiology at a high concentration? J. Microbiol. 2015, 53, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Donati, A.J.; Lee, H.-I.; Leveau, J.H.J.; Chang, W.-S. Effects of Indole-3-Acetic Acid on the Transcriptional Activities and Stress Tolerance of Bradyrhizobium japonicum. PLoS ONE 2013, 8, e76559. [Google Scholar] [CrossRef]
- Ahmed, M.; Stal, L.J.; Hasnain, S. Biofilm Formation and Indole-3-Acetic Acid Production by Two Rhizospheric Unicellular Cyanobacteria. J. Microbiol. Biotechnol. 2014, 24, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhang, C.; Mu, Y.; Shen, Q.; Feng, Y. Indole Affects Biofilm Formation in Bacteria. Indian J. Microbiol. 2010, 50, 362–368. [Google Scholar] [CrossRef]
- Pandit, A.; Adholeya, A.; Cahill, D.; Brau, L.; Kochar, M. Microbial biofilms in nature: Unlocking their potential for agricultural applications. J. Appl. Microbiol. 2020, 129, 199–211. [Google Scholar] [CrossRef]
- Baggs, E.L.; Monroe, J.G.; Thanki, A.S.; O’grady, R.; Schudoma, C.; Haerty, W.; Krasileva, K.V. Convergent Loss of an EDS1/PAD4 Signaling Pathway in Several Plant Lineages Reveals Coevolved Components of Plant Immunity and Drought Response. Plant Cell 2020, 32, 2158–2177. [Google Scholar] [CrossRef]
- Radulović, O.; Stanković, S.; Uzelac, B.; Tadić, V.; Trifunović-Momčilov, M.; Lozo, J.; Marković, M. Phenol Removal Capacity of the Common Duckweed (Lemna minor L.) and Six Phenol-Resistant Bacterial Strains from Its Rhizosphere: In Vitro Evaluation at High Phenol Concentrations. Plants 2020, 9, 599. [Google Scholar] [CrossRef]
- Radulovic, O.; Petrić, M.; Raspor, M.; Stanojević, O.; Janakiev, T.; Tadić, V.; Stanković, S. Culture-Dependent Analysis of 16S rRNA Sequences Associated with the Rhizosphere of Lemna minor and Assessment of Bacterial Phenol-Resistance: Plant/Bacteria System for Potential Bioremediation—Part II. Pol. J. Environ. Stud. 2018, 28, 811–822. [Google Scholar] [CrossRef]
- Girard, L.; Lood, C.; Höfte, M.; Vandamme, P.; Rokni-Zadeh, H.; van Noort, V.; Lavigne, R.; De Mot, R. The Ever-Expanding Pseudomonas Genus: Description of 43 New Species and Partition of the Pseudomonas putida Group. Microorganisms 2021, 9, 1766. [Google Scholar] [CrossRef]
- Vélez, J.M.B.; Martínez, J.G.; Ospina, J.T.; Agudelo, S.O. Bioremediation potential of Pseudomonas genus isolates from residual water, capable of tolerating lead through mechanisms of exopolysaccharide production and biosorption. Biotechnol. Rep. 2021, 32, e00685. [Google Scholar] [CrossRef] [PubMed]
- Qessaoui, R.; Bouharroud, R.; Furze, J.N.; El Aalaoui, M.; Akroud, H.; Amarraque, A.; Van Vaerenbergh, J.; Tahzima, R.; Mayad, E.H.; Chebli, B. Applications of New Rhizobacteria Pseudomonas Isolates in Agroecology via Fundamental Processes Complementing Plant Growth. Sci. Rep. 2019, 9, 12832. [Google Scholar] [CrossRef]
- Rieusset, L.; Rey, M.; Muller, D.; Vacheron, J.; Gerin, F.; Dubost, A.; Comte, G.; Prigent-Combaret, C. Secondary metabolites from plant-associated Pseudomonas are overproduced in biofilm. Microb. Biotechnol. 2020, 13, 1562–1580. [Google Scholar] [CrossRef] [PubMed]
- Mohite, B. Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 2013, 13, 638–649. [Google Scholar] [CrossRef]
- Utami, D.; Kawahata, A.; Sugawara, M.; Jog, R.N.; Miwa, K.; Morikawa, M. Effect of Exogenous General Plant Growth Regulators on the Growth of the Duckweed Lemna minor. Front. Chem. 2018, 6, 251. [Google Scholar] [CrossRef]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2009, 38, D750–D753. [Google Scholar] [CrossRef]
- Leveau, J.H.; Gerards, S. Discovery of a bacterial gene cluster for catabolism of the plant hormone indole 3-acetic acid. FEMS Microbiol. Ecol. 2008, 65, 238–250. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant growth promoting rhizobacteria isolated from halophytes and drought-tolerant plants: Genomic characterisation and exploration of phyto-beneficial traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Mellidou, I.; Ainalidou, A.; Papadopoulou, A.; Leontidou, K.; Genitsaris, S.; Karagiannis, E.; Van de Poel, B.; Karamanoli, K. Comparative Transcriptomics and Metabolomics Reveal an Intricate Priming Mechanism Involved in PGPR-Mediated Salt Tolerance in Tomato. Front. Plant Sci. 2021, 12, 713984. [Google Scholar] [CrossRef] [PubMed]
- Libbert, E.; Risch, H. Interactions between Plants and Epiphytic Bacteria Regarding Their Auxin Metabolism. V. Isolation and Identification of the IAA-Producing and Destroying Bacteria from Pea Plants. Physiol. Plant. 1969, 22, 51–58. [Google Scholar] [CrossRef]
- Liu, W.-H.; Chen, F.-F.; Wang, C.-E.; Fu, H.-H.; Fang, X.-Q.; Ye, J.-R.; Shi, J.-Y. Indole-3-Acetic Acid in Burkholderia pyrrocinia JK-SH007: Enzymatic Identification of the Indole-3-Acetamide Synthesis Pathway. Front. Microbiol. 2019, 10, 2559. [Google Scholar] [CrossRef] [PubMed]
- Donoso, R.; Leiva-Novoa, P.; Zúñiga, A.; Timmermann, T.; Recabarren-Gajardo, G.; González, B. Biochemical and Genetic Bases of Indole-3-Acetic Acid (Auxin Phytohormone) Degradation by the Plant-Growth-Promoting Rhizobacterium Paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 2017, 83, e01991-16. [Google Scholar] [CrossRef] [PubMed]
- Rosendahl, L.; Jochimsen, B.U. In vitro indole-3-acetic acid uptake in symbiosomes from soybean (Glycine max L.) root nodules. Symbiosis 1995, 19, 99–110. [Google Scholar]
- Torres, D.; Mongiardini, E.; Donadío, F.; Donoso, R.; Recabarren-Gajardo, G.; Gualpa, J.; Spaepen, S.; Defez, R.; Lopez, G.; Bianco, C.; et al. Molecular and physiological analysis of indole-3-acetic acid degradation in Bradyrhizobium japonicum E109. Res. Microbiol. 2021, 172, 103814. [Google Scholar] [CrossRef]
- Harwood, C.S.; Rivelli, M.; Ornston, L.N. Aromatic acids are chemoattractants for Pseudomonas putida. J. Bacteriol. 1984, 160, 622–628. [Google Scholar] [CrossRef]
- Scott, J.C.; Greenhut, I.V.; Leveau, J.H.J. Functional Characterization of the Bacterial iac Genes for Degradation of the Plant Hormone Indole-3-Acetic Acid. J. Chem. Ecol. 2013, 39, 942–951. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
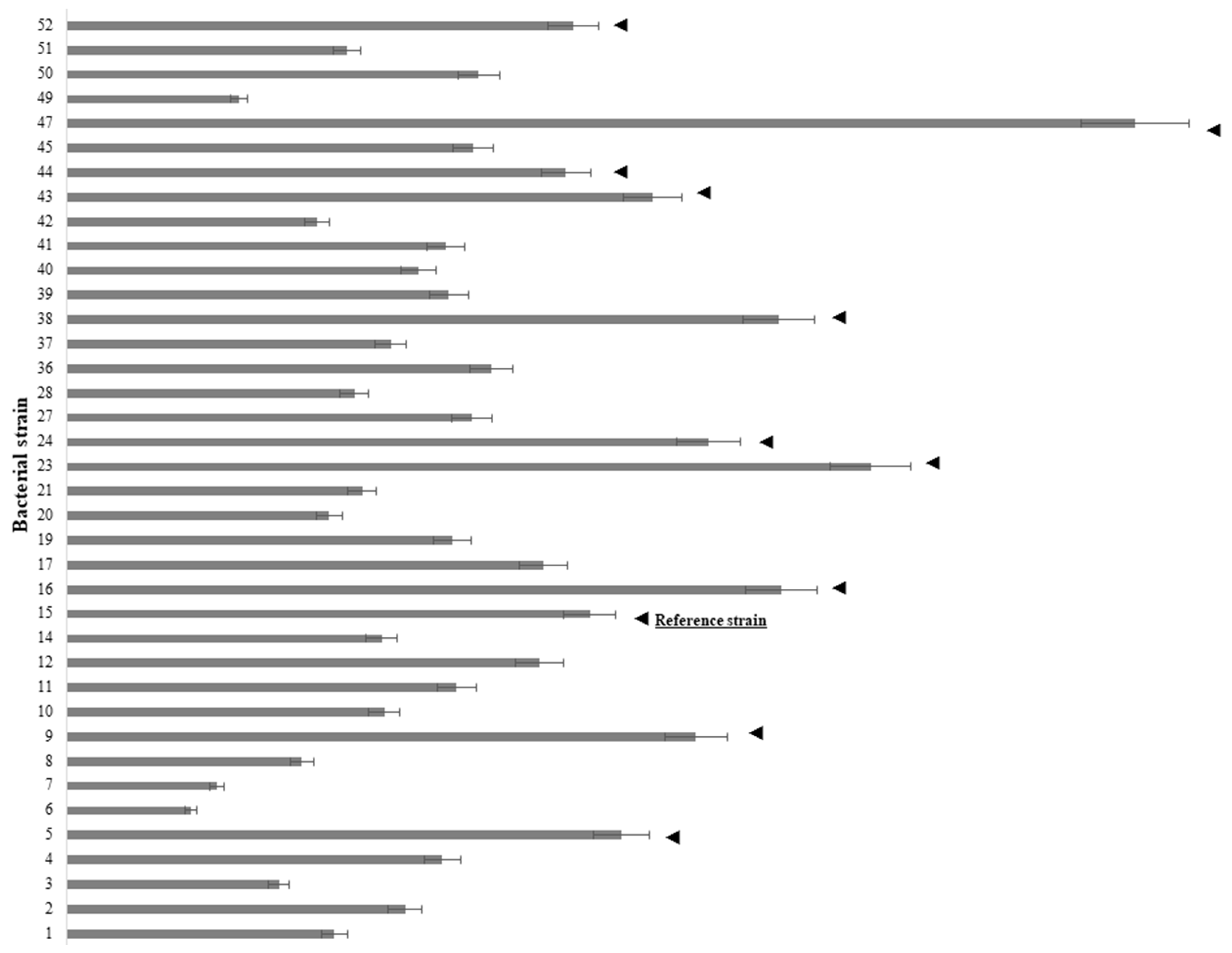



| Annotation | Strain | Species | Accession Number |
|---|---|---|---|
| 15 | D1-104/3 | Pseudomonas oryzihabitans | MF526913 |
| 23 | A3-104/5 | Pseudomonas putida | MF526920 |
| 38 | C31-106/3 | Pseudomonas gessardii | MF526935 |
| 47 | C44-104/1 | Pseudomonas yamanorum | MF526941 |
| Strain | Generation Time (h) | Growth Rate (h−1) | Relative Yield (OD600 Units Per 1 mmol of IAA) |
|---|---|---|---|
| P. oryzihabitans D1-104/3 | 7.64 ± 0.08 | 0.09 ± 0.01 | 0.08 ± 0.02 |
| P. putida A3-104/5 | 5.60 ± 0.05 | 0.12 ± 0.02 | 0.05 ± 0.01 |
| P. gessardii C31-106/3 | N/A | N/A | N/A |
| P. yamanorum C44-104/1 | 6.58 ± 0.05 | 0.10 ± 0.01 | 0.11 ± 0.05 |
| Strain | IAA Content (mg g−1 DW) | IAA Content (mg OD600−1) |
|---|---|---|
| P. oryzihabitans D1-104/3 | 7.41 ± 0.05 | 2.774 ± 0.005 |
| P. putida A3-104/5 | 5.47 ± 0.07 | 1.936 ± 0.005 |
| P. gessardii C31-106/3 | 1.12 ± 0.06 | 0.386 ± 0.009 |
| P. yamanorum C44-104/1 | 1.16 ± 0.05 | 0.626 ± 0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popržen, T.; Nikolić, I.; Krstić-Milošević, D.; Uzelac, B.; Trifunović-Momčilov, M.; Marković, M.; Radulović, O. Characterization of the IAA-Producing and -Degrading Pseudomonas Strains Regulating Growth of the Common Duckweed (Lemna minor L.). Int. J. Mol. Sci. 2023, 24, 17207. https://doi.org/10.3390/ijms242417207
Popržen T, Nikolić I, Krstić-Milošević D, Uzelac B, Trifunović-Momčilov M, Marković M, Radulović O. Characterization of the IAA-Producing and -Degrading Pseudomonas Strains Regulating Growth of the Common Duckweed (Lemna minor L.). International Journal of Molecular Sciences. 2023; 24(24):17207. https://doi.org/10.3390/ijms242417207
Chicago/Turabian StylePopržen, Tatjana, Ivan Nikolić, Dijana Krstić-Milošević, Branka Uzelac, Milana Trifunović-Momčilov, Marija Marković, and Olga Radulović. 2023. "Characterization of the IAA-Producing and -Degrading Pseudomonas Strains Regulating Growth of the Common Duckweed (Lemna minor L.)" International Journal of Molecular Sciences 24, no. 24: 17207. https://doi.org/10.3390/ijms242417207







