Ephexin3/ARHGEF5 Together with Cell Migration Signaling Partners within the Tumor Microenvironment Define Prognostic Transcriptional Signatures in Multiple Cancer Types
Abstract
:1. Introduction
2. Results
2.1. Expression of Ephexins in Various Cancer Types
2.2. Coexpression Patterns of Ephexins among Themselves and with Markers of Tumor Stromal Cells
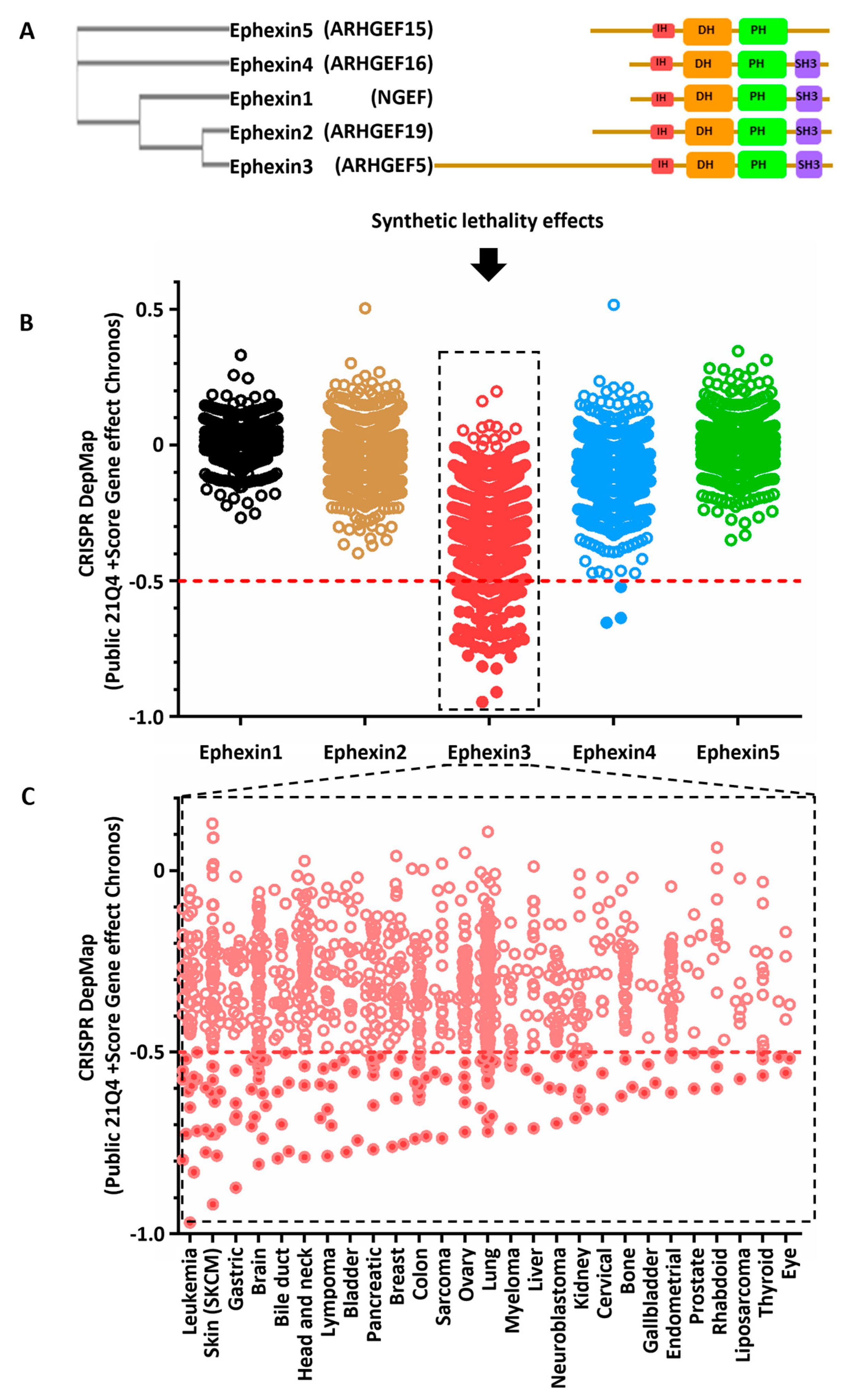
2.3. Cancer Cell Lines Sensitive to Ephexin3 Knock-Out
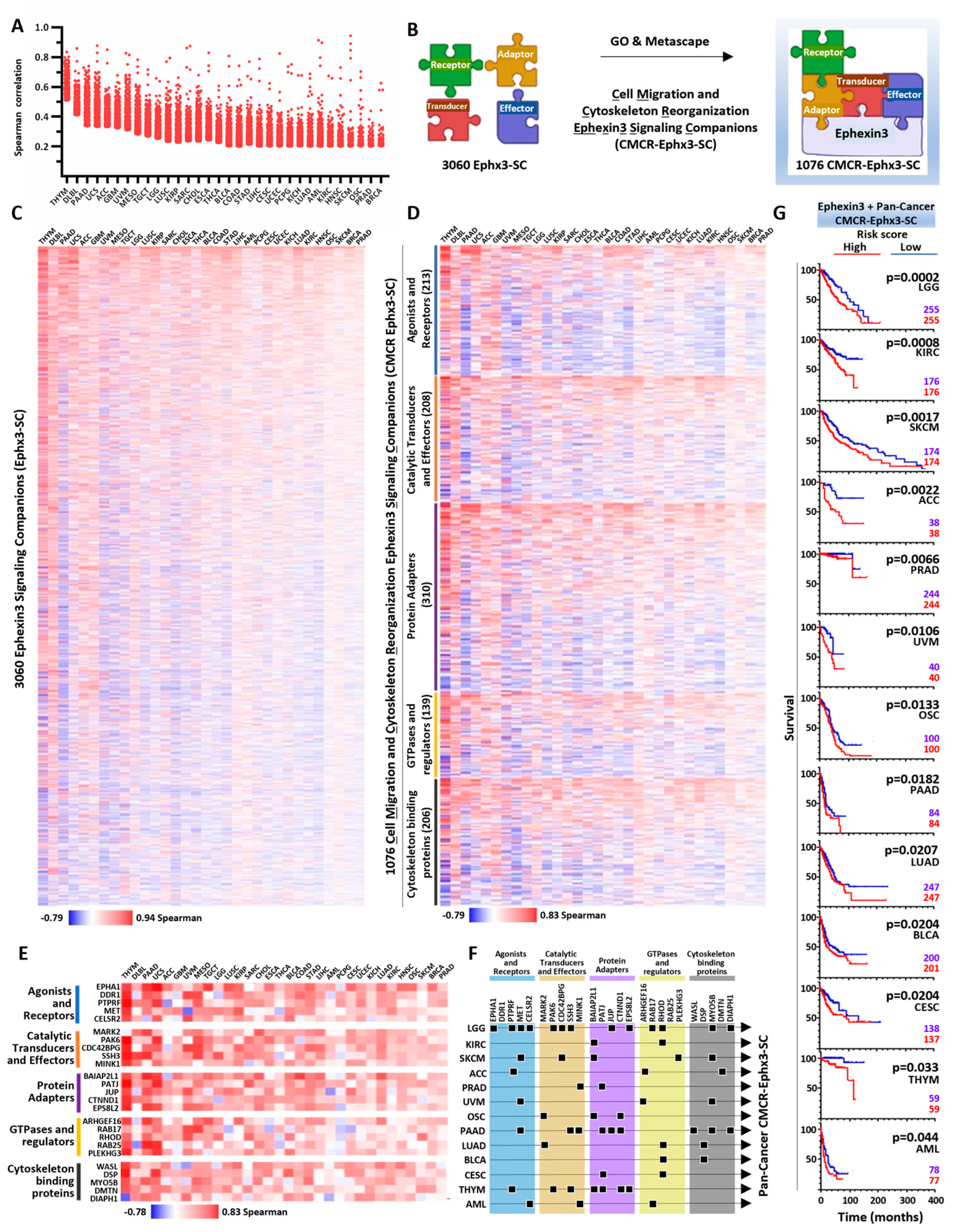
2.4. Ephexin3 Signaling Partners Linked to Cell Migration and Cytoskeleton Reorganization in 32 Cancer Types
2.5. PanCancer CMCR-Ephx3-SC Defining Transcriptional Signatures Statistically Linked to Shorter Patient Survival
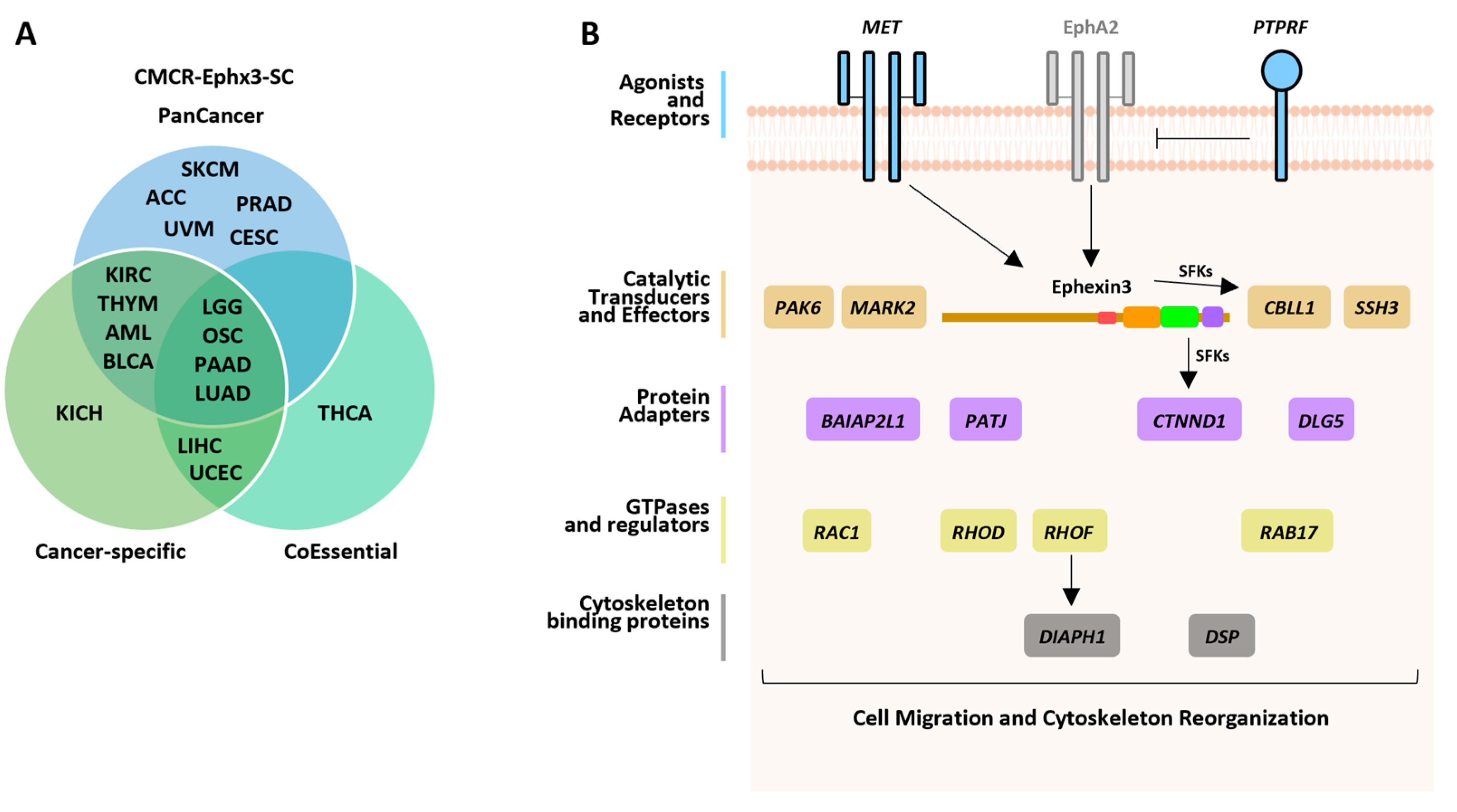
2.6. Cancer-Specific CMCR-Ephx3-SC Defining Transcriptional Signatures Statistically Linked to Shorter Patient Survival
2.7. CoEssential CMCR-Ephx3-SCs Defining Transcriptional Signatures Statistically Linked to Shorter Patient Survival
2.8. CMCR-Ephx3-SC Identified by Independent Data Mining Strategies
3. Discussion
4. Materials and Methods
4.1. Ephexins in TCGA Studies
4.2. Signaling Transcripts Coexpressed with Ephexins in TCGA Studies
4.3. Ephexins’ Correlation with Cell Type Markers
4.4. Ephexins’ Essentiality and CoEssential Partners
4.5. Ephexin3 Transcriptional Signatures
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, K.; Lee, S.A.; Park, D. Emerging Roles of Ephexins in Physiology and Disease. Cells 2019, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Villagrana, R.D.; Garcia-Jimenez, I.; Vazquez-Prado, J. Guanine nucleotide exchange factors for Rho GTPases (RhoGEFs) as oncogenic effectors and strategic therapeutic targets in metastatic cancer. Cell. Signal. 2023, 109, 110749. [Google Scholar] [CrossRef] [PubMed]
- Shamah, S.M.; Lin, M.Z.; Goldberg, J.L.; Estrach, S.; Sahin, M.; Hu, L.; Bazalakova, M.; Neve, R.L.; Corfas, G.; Debant, A.; et al. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 2001, 105, 233–244. [Google Scholar] [CrossRef]
- Janes, P.W.; Vail, M.E.; Ernst, M.; Scott, A.M. Eph Receptors in the Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 801–805. [Google Scholar] [CrossRef]
- Wang, Y.; Suzuki, H.; Yokoo, T.; Tada-Iida, K.; Kihara, R.; Miura, M.; Watanabe, K.; Sone, H.; Shimano, H.; Toyoshima, H.; et al. WGEF is a novel RhoGEF expressed in intestine, liver, heart, and kidney. Biochem. Biophys. Res. Commun. 2004, 324, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kumamoto, Y.; Wang, P.; Gan, X.; Lehmann, D.; Smrcka, A.V.; Cohn, L.; Iwasaki, A.; Li, L.; Wu, D. Regulation of immature dendritic cell migration by RhoA guanine nucleotide exchange factor Arhgef5. J. Biol. Chem. 2009, 284, 28599–28606. [Google Scholar] [CrossRef]
- Hiramoto-Yamaki, N.; Takeuchi, S.; Ueda, S.; Harada, K.; Fujimoto, S.; Negishi, M.; Katoh, H. Ephexin4 and EphA2 mediate cell migration through a RhoG-dependent mechanism. J. Cell Biol. 2010, 190, 461–477. [Google Scholar] [CrossRef]
- Kusuhara, S.; Fukushima, Y.; Fukuhara, S.; Jakt, L.M.; Okada, M.; Shimizu, Y.; Hata, M.; Nishida, K.; Negi, A.; Hirashima, M.; et al. Arhgef15 promotes retinal angiogenesis by mediating VEGF-induced Cdc42 activation and potentiating RhoJ inactivation in endothelial cells. PLoS ONE 2012, 7, e45858. [Google Scholar] [CrossRef]
- Kim, J.; Chang, I.Y.; You, H.J. Interactions between EGFR and EphA2 promote tumorigenesis through the action of Ephexin1. Cell Death Dis. 2022, 13, 528. [Google Scholar] [CrossRef]
- Kim, J.; Jeon, Y.J.; Lim, S.C.; Ryu, J.; Lee, J.H.; Chang, I.Y.; You, H.J. Akt-mediated Ephexin1-Ras interaction promotes oncogenic Ras signaling and colorectal and lung cancer cell proliferation. Cell Death Dis. 2021, 12, 1013. [Google Scholar] [CrossRef]
- Li, Y.; Ye, Z.; Chen, S.; Pan, Z.; Zhou, Q.; Li, Y.Z.; Shuai, W.D.; Kuang, C.M.; Peng, Q.H.; Shi, W.; et al. ARHGEF19 interacts with BRAF to activate MAPK signaling during the tumorigenesis of non-small cell lung cancer. Int. J. Cancer 2018, 142, 1379–1391. [Google Scholar] [CrossRef]
- He, P.; Wu, W.; Wang, H.; Liao, K.; Zhang, W.; Xiong, G.; Wu, F.; Meng, G.; Yang, K. Co-expression of Rho guanine nucleotide exchange factor 5 and Src associates with poor prognosis of patients with resected non-small cell lung cancer. Oncol. Rep. 2013, 30, 2864–2870. [Google Scholar] [CrossRef]
- Fukushima, H.; Yasumoto, M.; Ogasawara, S.; Akiba, J.; Kitasato, Y.; Nakayama, M.; Naito, Y.; Ishida, Y.; Okabe, Y.; Yasunaga, M.; et al. ARHGEF15 overexpression worsens the prognosis in patients with pancreatic ductal adenocarcinoma through enhancing the motility and proliferative activity of the cancer cells. Mol. Cancer 2016, 15, 32. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Villagrana, R.D.; Color-Aparicio, V.M.; Reyes-Cruz, G.; Vazquez-Prado, J. Protumoral bone marrow-derived cells migrate via Gbetagamma-dependent signaling pathways and exhibit a complex repertoire of RhoGEFs. J. Cell Commun. Signal. 2019, 13, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Onodera, Y.; Kuroiwa, M.; Nomimura, S.; Kubo, Y.; Nam, J.M.; Kajiwara, K.; Nada, S.; Oneyama, C.; Sabe, H.; et al. The Rho guanine nucleotide exchange factor ARHGEF5 promotes tumor malignancy via epithelial-mesenchymal transition. Oncogenesis 2016, 5, e258. [Google Scholar] [CrossRef]
- Wang, Z.; Kan, G.; Sheng, C.; Yao, C.; Mao, Y.; Chen, S. ARHGEF19 regulates MAPK/ERK signaling and promotes the progression of small cell lung cancer. Biochem. Biophys. Res. Commun. 2020, 533, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.M.; McGovern, E.S.; Catalano, G.; Fleming, T.P.; Miki, T. Expression cDNA cloning of a novel oncogene with sequence similarity to regulators of small GTP-binding proteins. Oncogene 1994, 9, 1057–1063. [Google Scholar] [PubMed]
- Kuroiwa, M.; Oneyama, C.; Nada, S.; Okada, M. The guanine nucleotide exchange factor Arhgef5 plays crucial roles in Src-induced podosome formation. J. Cell Sci. 2011, 124 Pt 10, 1726–1738. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, C.; Croucher, D.R.; Soliman, M.A.; St-Denis, N.; Pasculescu, A.; Taylor, L.; Tate, S.A.; Hardy, W.R.; Colwill, K.; et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 2013, 499, 166–171. [Google Scholar] [CrossRef]
- Beigbeder, A.; Chartier, F.J.M.; Bisson, N. MPZL1 forms a signalling complex with GRB2 adaptor and PTPN11 phosphatase in HER2-positive breast cancer cells. Sci. Rep. 2017, 7, 11514. [Google Scholar] [CrossRef]
- Guo, A.; Villen, J.; Kornhauser, J.; Lee, K.A.; Stokes, M.P.; Rikova, K.; Possemato, A.; Nardone, J.; Innocenti, G.; Wetzel, R.; et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc. Natl. Acad. Sci. USA 2008, 105, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Hiramoto-Yamaki, N.; Negishi, M.; Katoh, H. Ephexin4 and EphA2 mediate resistance to anoikis through RhoG and phosphatidylinositol 3-kinase. Exp. Cell Res. 2011, 317, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Xu, L.; Chen, L.; Wang, Y.; Jiang, H.; Yan, Y.; Luo, S.; Zhai, Z. FYN is required for ARHGEF16 to promote proliferation and migration in colon cancer cells. Cell Death Dis. 2020, 11, 652. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Wang, Y.; Xu, L.; Chen, L.; Cheng, M.; Shi, W.; Xiong, H.; Zalli, D.; Luo, S. GLI2 promotes cell proliferation and migration through transcriptional activation of ARHGEF16 in human glioma cells. J. Exp. Clin. Cancer Res. 2018, 37, 247. [Google Scholar] [CrossRef]
- Ding, L.; Bailey, M.H.; Porta-Pardo, E.; Thorsson, V.; Colaprico, A.; Bertrand, D.; Gibbs, D.L.; Weerasinghe, A.; Huang, K.L.; Tokheim, C.; et al. Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 2018, 173, 305–320.e10. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e10. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, Y.; Varambally, S.; Creighton, C.J. Molecular Correlates of Metastasis by Systematic Pan-Cancer Analysis Across The Cancer Genome Atlas. Mol. Cancer Res. 2019, 17, 476–487. [Google Scholar] [CrossRef]
- Akbani, R.; Ng, P.K.; Werner, H.M.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef]
- Boehm, J.S.; Garnett, M.J.; Adams, D.J.; Francies, H.E.; Golub, T.R.; Hahn, W.C.; Iorio, F.; McFarland, J.M.; Parts, L.; Vazquez, F. Cancer research needs a better map. Nature 2021, 589, 514–516. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Goncalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Lanczky, A.; Gyorffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Nguyen, B.; Fong, C.; Luthra, A.; Smith, S.A.; DiNatale, R.G.; Nandakumar, S.; Walch, H.; Chatila, W.K.; Madupuri, R.; Kundra, R.; et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 2022, 185, 563–575.e11. [Google Scholar] [CrossRef]
- Ganesh, K.; Massague, J. Targeting metastatic cancer. Nat. Med. 2021, 27, 34–44. [Google Scholar] [CrossRef]
- Weiss, F.; Lauffenburger, D.; Friedl, P. Towards targeting of shared mechanisms of cancer metastasis and therapy resistance. Nat. Rev. Cancer 2022, 22, 157–173. [Google Scholar] [CrossRef]
- Jansen, S.; Gosens, R.; Wieland, T.; Schmidt, M. Paving the Rho in cancer metastasis: Rho GTPases and beyond. Pharmacol. Ther. 2018, 183, 1–21. [Google Scholar] [CrossRef]
- Gillette, M.A.; Satpathy, S.; Cao, S.; Dhanasekaran, S.M.; Vasaikar, S.V.; Krug, K.; Petralia, F.; Li, Y.; Liang, W.W.; Reva, B.; et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020, 182, 200–225.e35. [Google Scholar] [CrossRef]
- He, P.; Wu, W.; Yang, K.; Tan, D.; Tang, M.; Liu, H.; Wu, T.; Zhang, S.; Wang, H. Rho Guanine Nucleotide Exchange Factor 5 Increases Lung Cancer Cell Tumorigenesis via MMP-2 and Cyclin D1 Upregulation. Mol. Cancer Ther. 2015, 14, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Debily, M.A.; Camarca, A.; Ciullo, M.; Mayer, C.; El Marhomy, S.; Ba, I.; Jalil, A.; Anzisi, A.; Guardiola, J.; Piatier-Tonneau, D. Expression and molecular characterization of alternative transcripts of the ARHGEF5/TIM oncogene specific for human breast cancer. Hum. Mol. Genet. 2004, 13, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Chang, S.W.; Tatsumoto, T.; Chan, A.M.; Miki, T. TIM, a Dbl-related protein, regulates cell shape and cytoskeletal organization in a Rho-dependent manner. Cell. Signal. 2005, 17, 461–471. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, L.; Wang, C.; Zhu, J. Double inhibition and activation mechanisms of Ephexin family RhoGEFs. Proc. Natl. Acad. Sci. USA 2021, 118, e2024465118. [Google Scholar] [CrossRef]
- Feng, X.; Arang, N.; Rigiracciolo, D.C.; Lee, J.S.; Yeerna, H.; Wang, Z.; Lubrano, S.; Kishore, A.; Pachter, J.A.; Konig, G.M.; et al. A Platform of Synthetic Lethal Gene Interaction Networks Reveals that the GNAQ Uveal Melanoma Oncogene Controls the Hippo Pathway through FAK. Cancer Cell 2019, 35, 457–472.e5. [Google Scholar] [CrossRef]
- Feng, X.; Degese, M.S.; Iglesias-Bartolome, R.; Vaque, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef]
- Crosas-Molist, E.; Samain, R.; Kohlhammer, L.; Orgaz, J.L.; George, S.L.; Maiques, O.; Barcelo, J.; Sanz-Moreno, V. Rho GTPase signaling in cancer progression and dissemination. Physiol. Rev. 2022, 102, 455–510. [Google Scholar] [CrossRef] [PubMed]
- Yu, O.M.; Brown, J.H. G Protein-Coupled Receptor and RhoA-Stimulated Transcriptional Responses: Links to Inflammation, Differentiation, and Cell Proliferation. Mol. Pharmacol. 2015, 88, 171–180. [Google Scholar] [CrossRef]
- Olson, E.N.; Nordheim, A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010, 11, 353–365. [Google Scholar] [CrossRef]
- Lionarons, D.A.; Hancock, D.C.; Rana, S.; East, P.; Moore, C.; Murillo, M.M.; Carvalho, J.; Spencer-Dene, B.; Herbert, E.; Stamp, G.; et al. RAC1(P29S) Induces a Mesenchymal Phenotypic Switch via Serum Response Factor to Promote Melanoma Development and Therapy Resistance. Cancer Cell 2019, 36, 68–83.e9. [Google Scholar] [CrossRef]
- Franklin, J.M.; Wu, Z.M.; Guan, K.L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat. Rev. Cancer 2023, 23, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Thrash, H.L.; Pendergast, A.M. Multi-Functional Regulation by YAP/TAZ Signaling Networks in Tumor Progression and Metastasis. Cancers 2023, 15, 4701. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Saito, A.; Nagase, T. YAP/TAZ Signaling as a Molecular Link between Fibrosis and Cancer. Int. J. Mol. Sci. 2018, 19, 3674. [Google Scholar] [CrossRef] [PubMed]
- Mihira, H.; Suzuki, H.I.; Akatsu, Y.; Yoshimatsu, Y.; Igarashi, T.; Miyazono, K.; Watabe, T. TGF-beta-induced mesenchymal transition of MS-1 endothelial cells requires Smad-dependent cooperative activation of Rho signals and MRTF-A. J. Biochem. 2012, 151, 145–156. [Google Scholar] [CrossRef]
- Robatel, S.; Schenk, M. Current Limitations and Novel Perspectives in Pancreatic Cancer Treatment. Cancers 2022, 14, 985. [Google Scholar] [CrossRef]
- Fard, D.; Giraudo, E.; Tamagnone, L. Mind the (guidance) signals! Translational relevance of semaphorins, plexins, and neuropilins in pancreatic cancer. Trends Mol. Med. 2023, 29, 817–829. [Google Scholar] [CrossRef]
- Leung, A.W.; de Silva, T.; Bally, M.B.; Lockwood, W.W. Synthetic lethality in lung cancer and translation to clinical therapies. Mol. Cancer 2016, 15, 61. [Google Scholar] [CrossRef]
- Fu, J.; Su, X.; Li, Z.; Deng, L.; Liu, X.; Feng, X.; Peng, J. HGF/c-MET pathway in cancer: From molecular characterization to clinical evidence. Oncogene 2021, 40, 4625–4651. [Google Scholar] [CrossRef]
- Cooke, M.; Kreider-Letterman, G.; Baker, M.J.; Zhang, S.; Sullivan, N.T.; Eruslanov, E.; Abba, M.C.; Goicoechea, S.M.; Garcia-Mata, R.; Kazanietz, M.G. FARP1, ARHGEF39, and TIAM2 are essential receptor tyrosine kinase effectors for Rac1-dependent cell motility in human lung adenocarcinoma. Cell Rep. 2021, 37, 109905. [Google Scholar] [CrossRef]
- Delitto, D.; Zhang, D.; Han, S.; Black, B.S.; Knowlton, A.E.; Vlada, A.C.; Sarosi, G.A.; Behrns, K.E.; Thomas, R.M.; Lu, X.; et al. Nicotine Reduces Survival via Augmentation of Paracrine HGF-MET Signaling in the Pancreatic Cancer Microenvironment. Clin. Cancer Res. 2016, 22, 1787–1799. [Google Scholar] [CrossRef]
- Blom, M.; Reis, K.; Heldin, J.; Kreuger, J.; Aspenstrom, P. The atypical Rho GTPase RhoD is a regulator of actin cytoskeleton dynamics and directed cell migration. Exp. Cell Res. 2017, 352, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Durkin, C.H.; Leite, F.; Cordeiro, J.V.; Handa, Y.; Arakawa, Y.; Valderrama, F.; Way, M. RhoD Inhibits RhoC-ROCK-Dependent Cell Contraction via PAK6. Dev. Cell 2017, 41, 315–329.e7. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Pellegrin, S.; Scott, A.; Mellor, H. The small GTPase Rif is an alternative trigger for the formation of actin stress fibers in epithelial cells. J. Cell Sci. 2010, 123 Pt 8, 1247–1252. [Google Scholar] [CrossRef]
- Morse, E.M.; Sun, X.; Olberding, J.R.; Ha, B.H.; Boggon, T.J.; Calderwood, D.A. PAK6 targets to cell-cell adhesions through its N-terminus in a Cdc42-dependent manner to drive epithelial colony escape. J. Cell Sci. 2016, 129, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Fram, S.; King, H.; Sacks, D.B.; Wells, C.M. A PAK6-IQGAP1 complex promotes disassembly of cell-cell adhesions. Cell Mol. Life Sci. 2014, 71, 2759–2773. [Google Scholar] [CrossRef]
- Watanabe, N.; Kato, T.; Fujita, A.; Ishizaki, T.; Narumiya, S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1999, 1, 136–143. [Google Scholar] [CrossRef]
- Narumiya, S.; Tanji, M.; Ishizaki, T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009, 28, 65–76. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juan-Guadarrama, D.G.; Beltrán-Navarro, Y.M.; Reyes-Cruz, G.; Vázquez-Prado, J. Ephexin3/ARHGEF5 Together with Cell Migration Signaling Partners within the Tumor Microenvironment Define Prognostic Transcriptional Signatures in Multiple Cancer Types. Int. J. Mol. Sci. 2023, 24, 16427. https://doi.org/10.3390/ijms242216427
Juan-Guadarrama DG, Beltrán-Navarro YM, Reyes-Cruz G, Vázquez-Prado J. Ephexin3/ARHGEF5 Together with Cell Migration Signaling Partners within the Tumor Microenvironment Define Prognostic Transcriptional Signatures in Multiple Cancer Types. International Journal of Molecular Sciences. 2023; 24(22):16427. https://doi.org/10.3390/ijms242216427
Chicago/Turabian StyleJuan-Guadarrama, Dante Gustavo, Yarely Mabell Beltrán-Navarro, Guadalupe Reyes-Cruz, and José Vázquez-Prado. 2023. "Ephexin3/ARHGEF5 Together with Cell Migration Signaling Partners within the Tumor Microenvironment Define Prognostic Transcriptional Signatures in Multiple Cancer Types" International Journal of Molecular Sciences 24, no. 22: 16427. https://doi.org/10.3390/ijms242216427





