Biophysical Insights into the Antitumoral Activity of Crotalicidin against Breast Cancer Model Membranes
Abstract
:1. Introduction
2. Results
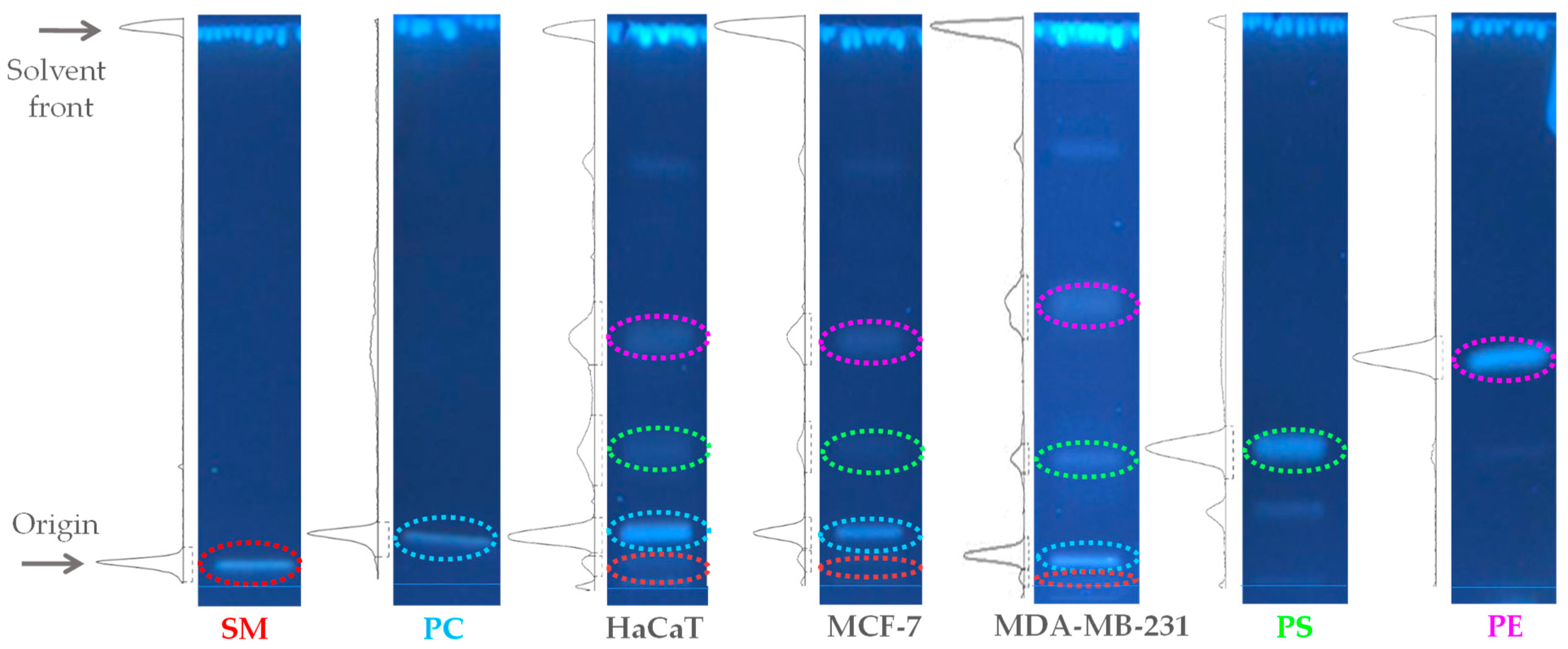
2.1. Quantification of the Total Lipid Extracts of MDA-MB-231, MCF-7, and HaCaT Cell Membranes
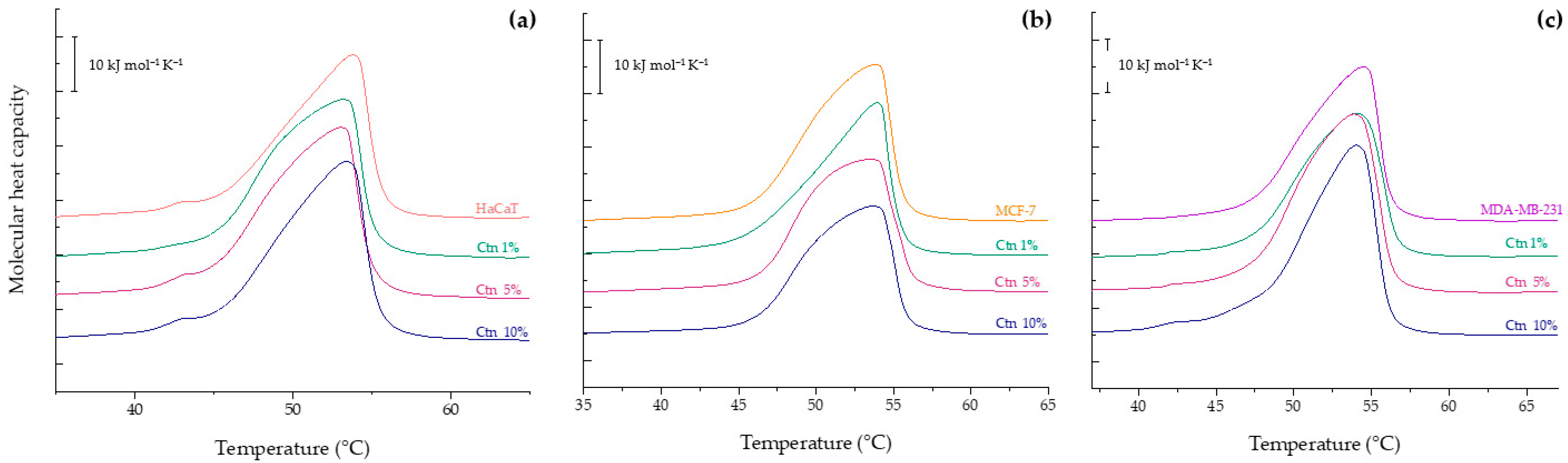
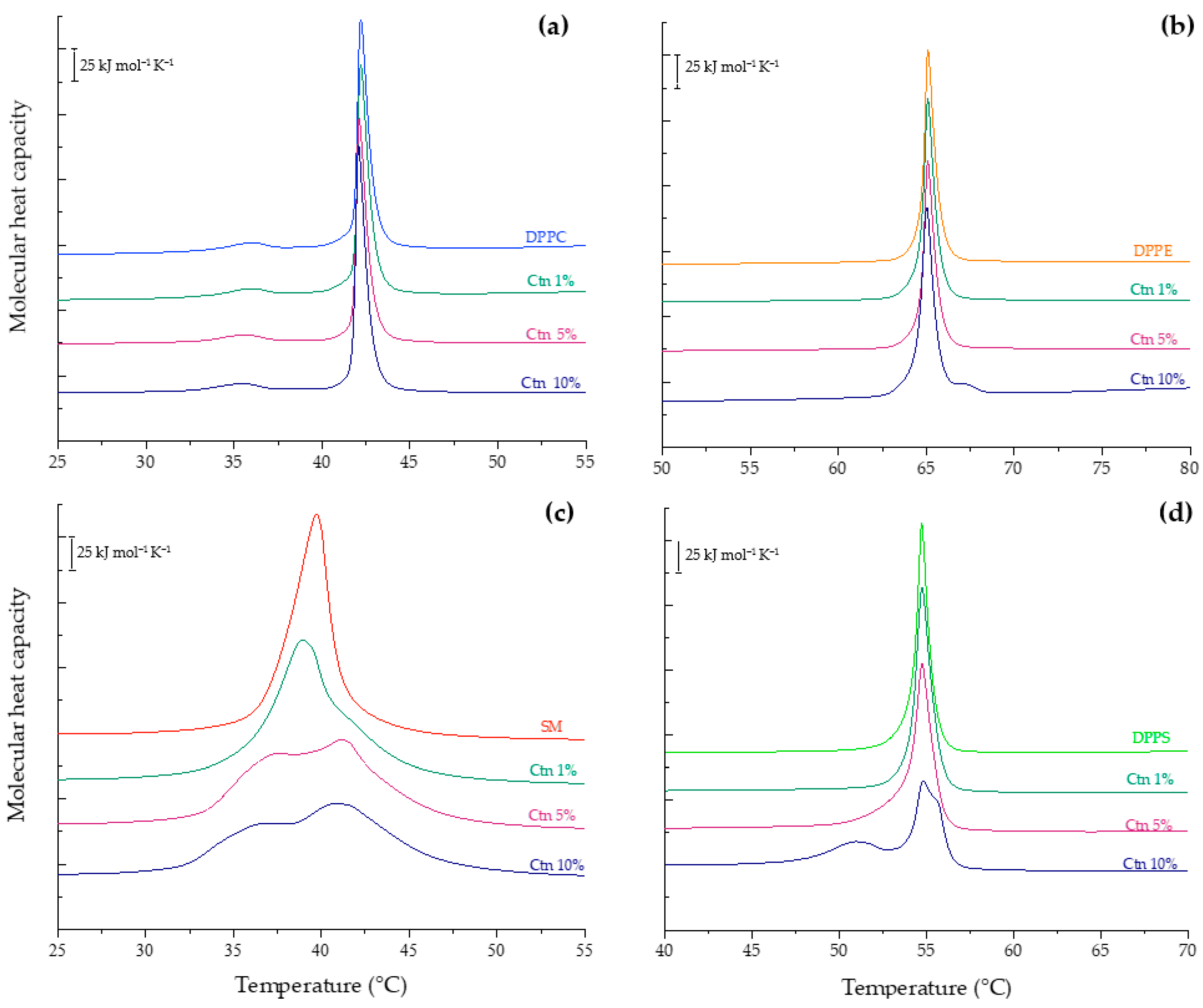
2.2. Differential Scanning Calorimetry Experiments
2.2.1. Evaluation of the Effect of Ctn on HaCaT, MCF-7 and MDA-MB-231 Model Membranes
2.2.2. Evaluation of the Effect of Ctn on Individual Lipid Systems
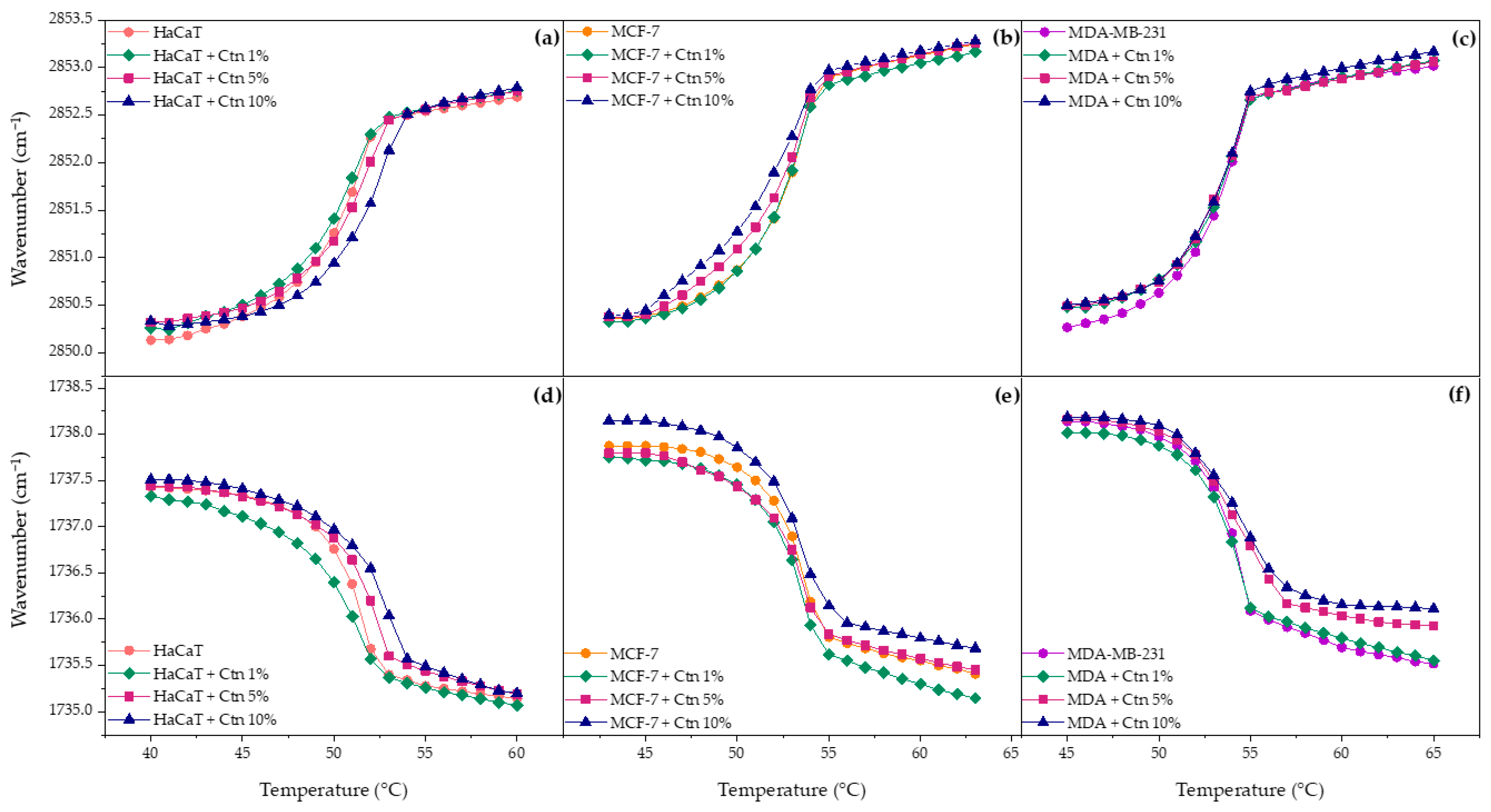
2.3. Phase Transition Measurements
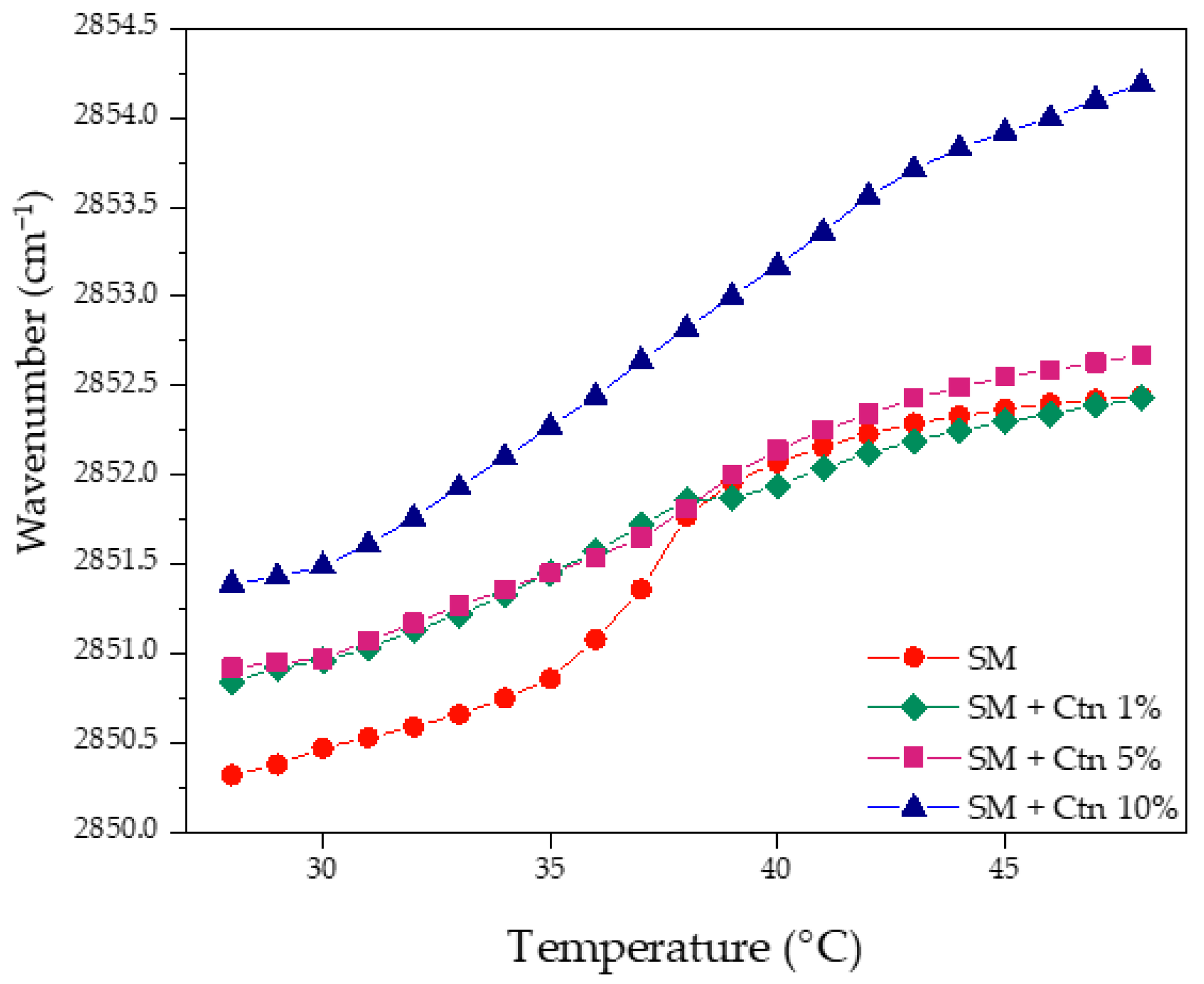
2.3.1. Multi-Component Lipid Systems Using FT-IR
2.3.2. Individual Pure Lipid Systems Using FT-IR
2.4. Peptide Structure Prediction Methods
2.4.1. De Novo Structure Prediction
2.4.2. BPROT1 Method
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture Conditions and Total Lipid Extraction
4.3. High-Performance Thin-Layer Chromatography (HPTLC)
4.4. Differential Scanning Calorimetry (DSC)
4.5. Fourier-Transform Infrared Spectroscopy (FT-IR)
4.6. Peptide Structure Prediction Methods
4.6.1. De Novo Structure Prediction of Ctn
4.6.2. Conformational Study of Ctn
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, G.; Huang, Y.; Feng, Q.; Chen, Y. Tryptophan as a probe to study the anticancer mechanism of action and specificity of α-helical anticancer peptides. Molecules 2014, 19, 12224–12241. [Google Scholar] [CrossRef] [PubMed]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [PubMed]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.N.; Soares, A.M.; Da Silva, S.L. Why to Study Peptides from Venomous and Poisonous Animals? Int. J. Pept. Res. Ther. 2023, 29, 76. [Google Scholar] [CrossRef]
- Falcao, C.; de La Torre, B.; Pérez-Peinado, C.; Barron, A.; Andreu, D.; Rádis-Baptista, G. Vipericidins: A novel family of cathelicidin-related peptides from the venom gland of South American pit vipers. Amino Acids 2014, 46, 2561–2571. [Google Scholar] [CrossRef]
- Falcao, C.B.; Pérez-Peinado, C.; de la Torre, B.G.; Mayol, X.; Zamora-Carreras, H.c.; Jiménez, M.A.n.; Rádis-Baptista, G.; Andreu, D. Structural dissection of crotalicidin, a rattlesnake venom cathelicidin, retrieves a fragment with antimicrobial and antitumor activity. J. Med. Chem. 2015, 58, 8553–8563. [Google Scholar] [CrossRef]
- Cavalcante, C.S.P.; de Aguiar, F.L.L.; Fontenelle, R.O.S.; de Menezes, R.R.d.P.P.B.; Martins, A.M.C.; Falcão, C.B.; Andreu, D.; Rádis-Baptista, G. Insights into the candidacidal mechanism of Ctn[15–34]—A carboxyl-terminal, crotalicidin-derived peptide related to cathelicidins. J. Med. Microbiol. 2018, 67, 129–138. [Google Scholar] [CrossRef]
- Bandeira, I.; Bandeira-Lima, D.; Mello, C.; Pereira, T.; De Menezes, R.; Sampaio, T.; Martins, A. Antichagasic effect of crotalicidin, a cathelicidin-like vipericidin, found in Crotalus durissus terrificus rattlesnake’s venom gland. Parasitology 2017, 8, 1059–1064. [Google Scholar] [CrossRef]
- Pérez-Peinado, C.; Dias, S.A.; Domingues, M.M.; Benfield, A.H.; Freire, J.M.; Rádis-Baptista, G.; Gaspar, D.; Castanho, M.A.; Craik, D.J.; Henriques, S.T. Mechanisms of bacterial membrane permeabilization by crotalicidin (Ctn) and its fragment Ctn (15–34), antimicrobial peptides from rattlesnake venom. J. Biol. Chem. 2018, 293, 1536–1549. [Google Scholar] [CrossRef]
- Trinidad-Calderón, P.A.; Varela-Chinchilla, C.D.; García-Lara, S. Natural Peptides Inducing Cancer Cell Death: Mechanisms and Properties of Specific Candidates for Cancer Therapeutics. Molecules 2021, 26, 7453. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Weibel, D.B. Organization and function of anionic phospholipids in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 4255–4267. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.W.; Garris, C.; Pfirschke, C.; Rickelt, S.; Arlauckas, S.; Siwicki, M.; Kohler, R.H.; Weissleder, R.; Sundvold-Gjerstad, V.; Sveinbjørnsson, B.; et al. LTX-315 sequentially promotes lymphocyte-independent and lymphocyte-dependent antitumor effects. Cell Stress 2019, 3, 348–360. [Google Scholar] [CrossRef]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef]
- Camilio, K.A.; Rekdal, Ø.; Sveinbjörnsson, B. LTX-315 (Oncopore™) A short synthetic anticancer peptide and novel immunotherapeutic agent. Oncoimmunology 2014, 3, e29181. [Google Scholar] [CrossRef]
- Pinault, M.; Guimaraes, C.; Dumas, J.-F.; Servais, S.; Chevalier, S.; Besson, P.; Goupille, C. A 1D High Performance Thin Layer Chromatography Method Validated to Quantify Phospholipids Including Cardiolipin and Monolysocardiolipin from Biological Samples. Eur. J. Lipid Sci. Technol. 2020, 122, 1900240. [Google Scholar] [CrossRef]
- Dória, M.L.; Cotrim, Z.; Macedo, B.; Simões, C.; Domingues, P.; Helguero, L.; Domingues, M.R. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 2011, 133, 635–648. [Google Scholar] [CrossRef]
- Riske, K.A.; Barroso, R.P.; Vequi-Suplicy, C.C.; Germano, R.; Henriques, V.B.; Lamy, M.T. Lipid bilayer pre-transition as the beginning of the melting process. Biochim. Biophys. Acta Biomembr. 2009, 1788, 954–963. [Google Scholar] [CrossRef]
- Çetinel, Z.Ö.; Bilge, D. The effects of miltefosine on the structure and dynamics of DPPC and DPPS liposomes mimicking normal and cancer cell membranes: FTIR and DSC studies. J. Mol. Liq. 2022, 356, 119041. [Google Scholar] [CrossRef]
- Lewis, R.N.; McElhaney, R.N. Calorimetric and spectroscopic studies of the polymorphic phase behavior of a homologous series of n-saturated 1,2-diacyl phosphatidylethanolamines. Biophys. J. 1993, 64, 1081–1096. [Google Scholar] [CrossRef] [PubMed]
- Klaiss-Luna, M.C.; Jemioła-Rzemińska, M.; Strzałka, K.; Manrique-Moreno, M. Understanding the Biophysical Interaction of LTX-315 with Tumoral Model Membranes. Int. J. Mol. Sci. 2023, 24, 581. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Falanga, A.; Cantisani, M.; Vitiello, M.; Morelli, G.; Galdiero, M. Peptide-lipid interactions: Experiments and applications. Int. J. Mol. Sci. 2013, 14, 18758–18789. [Google Scholar] [CrossRef] [PubMed]
- Marín-Medina, N.; Ramírez, D.A.; Trier, S.; Leidy, C. Mechanical properties that influence antimicrobial peptide activity in lipid membranes. Appl. Microbiol. Biotechnol. 2016, 100, 10251–10263. [Google Scholar] [CrossRef] [PubMed]
- Mai, X.-T.; Huang, J.; Tan, J.; Huang, Y.; Chen, Y. Effects and mechanisms of the secondary structure on the antimicrobial activity and specificity of antimicrobial peptides. J. Pept. Sci. 2015, 21, 561–568. [Google Scholar] [CrossRef]
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theor. Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef]
- Symons, J.L.; Cho, K.-J.; Chang, J.T.; Du, G.; Waxham, M.N.; Hancock, J.F.; Levental, I.; Levental, K.R. Lipidomic atlas of mammalian cell membranes reveals hierarchical variation induced by culture conditions, subcellular membranes, and cell lineages. Soft Matter 2021, 17, 288–297. [Google Scholar] [CrossRef]
- Shah, T.; Krishnamachary, B.; Wildes, F.; Wijnen, J.P.; Glunde, K.; Bhujwalla, Z.M. Molecular causes of elevated phosphoethanolamine in breast and pancreatic cancer cells. NMR Biomed. 2018, 31, e3936. [Google Scholar] [CrossRef]
- Kim, H.Y.; Lee, K.M.; Kim, S.H.; Kwon, Y.J.; Chun, Y.J.; Choi, H.K. Comparative metabolic and lipidomic profiling of human breast cancer cells with different metastatic potentials. Oncotarget 2016, 7, 67111–67128. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.C.; Ribeiro, D.; Nunes, C.; Reis, S. Biophysics in cancer: The relevance of drug-membrane interaction studies. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2231–2244. [Google Scholar] [CrossRef] [PubMed]
- Rivel, T.; Ramseyer, C.; Yesylevskyy, S. The asymmetry of plasma membranes and their cholesterol content influence the uptake of cisplatin. Sci. Rep. 2019, 9, 5627. [Google Scholar] [CrossRef] [PubMed]
- Pašalić, L.; Jakas, A.; Pem, B.; Bakarić, D. Adsorption/Desorption of Cationic-Hydrophobic Peptides on Zwitterionic Lipid Bilayer Is Associated with the Possibility of Proton Transfer. Antibiotics 2023, 12, 1216. [Google Scholar] [CrossRef]
- Powers, J.P.; Tan, A.; Ramamoorthy, A.; Hancock, R.E. Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry 2005, 44, 15504–15513. [Google Scholar] [CrossRef]
- Sani, M.-A.; Separovic, F. How Membrane-Active Peptides Get into Lipid Membranes. Acc. Chem. Res. 2016, 49, 1130–1138. [Google Scholar] [CrossRef]
- Pignatello, R. Drug-Biomembrane Interaction Studies: The Application of Calorimetric Techniques; Elsevier: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Mbadugha, B.N.A.; Menger, F.M. Sugar/Steroid/Sugar Conjugates: Sensitivity of Lipid Binding to Sugar Structure. Org. Lett. 2003, 5, 4041–4044. [Google Scholar] [CrossRef]
- Camilio, K.A.; Berge, G.; Ravuri, C.S.; Rekdal, Ø.; Sveinbjørnsson, B. Complete regression and systemic protective immune responses obtained in B16 melanomas after treatment with LTX-315. Cancer Immunol. Immunother. 2014, 63, 601–613. [Google Scholar] [CrossRef]
- Niemela, P.; Hyvönen, M.T.; Vattulainen, I. Structure and Dynamics of Sphingomyelin Bilayer: Insight Gained through Systematic Comparison to Phosphatidylcholine. Biophys. J. 2004, 87, 2976–2989. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Haney, E.; Nazmi, K.; Lau, F.; Bolscher, J.; Vogel, H. Novel lactoferrampin antimicrobial peptides derived from human lactoferrin. Biochimie 2009, 91, 141–154. [Google Scholar] [CrossRef]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef] [PubMed]
- Ali, O.; Szabó, A. Review of Eukaryote Cellular Membrane Lipid Composition, with Special Attention to the Fatty Acids. Int. J. Mol. Sci. 2023, 24, 15693. [Google Scholar] [CrossRef]
- Cseke, L.J.; Kirakosyan, A.; Kaufman, P.B.; Westfall, M.V. Handbook of Molecular and Cellular Methods in Biology and Medicine; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Klaiss-Luna, M.C.; Manrique-Moreno, M. Infrared Spectroscopic Study of Multi-Component Lipid Systems: A Closer Approximation to Biological Membrane Fluidity. Membranes 2022, 12, 534. [Google Scholar] [CrossRef] [PubMed]


| Heating | |||
|---|---|---|---|
| T [°C] | ΔH [kJ mol−1] | ΔS [kJ mol−1 K−1] | |
| HaCaT * | 42.93 and 53.79 | 36.94 | 0.11 |
| +1 mol% Ctn | 53.21 | 39.36 | 0.12 |
| +5 mol% Ctn * | 43.21 and 53.05 | 41.50 | 0.13 |
| +10 mol% Ctn * | 43.16 and 53.39 | 41.99 | 0.13 |
| MCF-7 | 53.86 | 38.51 | 0.12 |
| +1 mol% Ctn | 53.94 | 34.14 | 0.10 |
| +5 mol% Ctn | 53.51 | 35.32 | 0.11 |
| +10 mol% Ctn | 53.67 | 33.48 | 0.10 |
| MDA-MB-231 | 54.47 | 41.47 | 0.13 |
| +1 mol% Ctn * | 42.07 and 54.01 | 42.77 | 0.13 |
| +5 mol% Ctn * | 42.20 and 53.91 | 40.24 | 0.12 |
| +10 mol% Ctn * | 42.50 and 54.03 | 39.64 | 0.12 |
| Heating | |||
|---|---|---|---|
| T [°C] | ΔH [kJ mol−1] | ΔS [kJ mol−1 K−1] | |
| DPPC | 42.20 35.84 (#) | 32.37 2.84 | 0.10 0.01 |
| +1 mol% Ctn | 42.11 35.26 | 28.86 1.65 | 0.09 0.01 |
| +5 mol% Ctn | 42.12 35.57 | 29.05 2.38 | 0.09 0.01 |
| +10 mol% Ctn | 42.08 35.40 | 30.92 2.39 | 0.10 0.01 |
| DPPE | 65.04 | 35.60 | 0.11 |
| +1 mol% Ctn | 65.07 | 33.61 | 0.10 |
| +5 mol% Ctn | 65.07 | 30.46 | 0.09 |
| +10 mol% Ctn | 65.02 | 36.32 | 0.11 |
| SM | 39.73 | 42.00 | 0.13 |
| +1 mol% Ctn | 38.93 | 38.93 | 0.12 |
| +5 mol% Ctn * | 37.5 and 41.13 | 38.27 | 0.12 |
| +10 mol% Ctn * | 36.82 and 40.84 | 36.15 | 0.12 |
| DPPS | 54.69 | 35.48 | 0.11 |
| +1 mol% Ctn | 54.73 | 36.62 | 0.11 |
| +5 mol% Ctn | 54.72 | 38.28 | 0.12 |
| +10 mol% Ctn * | 50.93, 54.78 and 55.49 | 34.29 | 0.10 |
| System | Helical Increase (%) * |
|---|---|
| POPC | 3.8 |
| HaCaT | 12.5 |
| MCF-7 | 10.2 |
| MDA-MB-231 | 13.7 |
| POPS | 46.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klaiss-Luna, M.C.; Giraldo-Lorza, J.M.; Jemioła-Rzemińska, M.; Strzałka, K.; Manrique-Moreno, M. Biophysical Insights into the Antitumoral Activity of Crotalicidin against Breast Cancer Model Membranes. Int. J. Mol. Sci. 2023, 24, 16226. https://doi.org/10.3390/ijms242216226
Klaiss-Luna MC, Giraldo-Lorza JM, Jemioła-Rzemińska M, Strzałka K, Manrique-Moreno M. Biophysical Insights into the Antitumoral Activity of Crotalicidin against Breast Cancer Model Membranes. International Journal of Molecular Sciences. 2023; 24(22):16226. https://doi.org/10.3390/ijms242216226
Chicago/Turabian StyleKlaiss-Luna, Maria C., Juan M. Giraldo-Lorza, Małgorzata Jemioła-Rzemińska, Kazimierz Strzałka, and Marcela Manrique-Moreno. 2023. "Biophysical Insights into the Antitumoral Activity of Crotalicidin against Breast Cancer Model Membranes" International Journal of Molecular Sciences 24, no. 22: 16226. https://doi.org/10.3390/ijms242216226






