Proteomic Determinants of Variation in Cholesterol Efflux: Observations from the Dallas Heart Study
Abstract
:1. Introduction
2. Results
2.1. Study Population Demographics and Clinical Characteristics
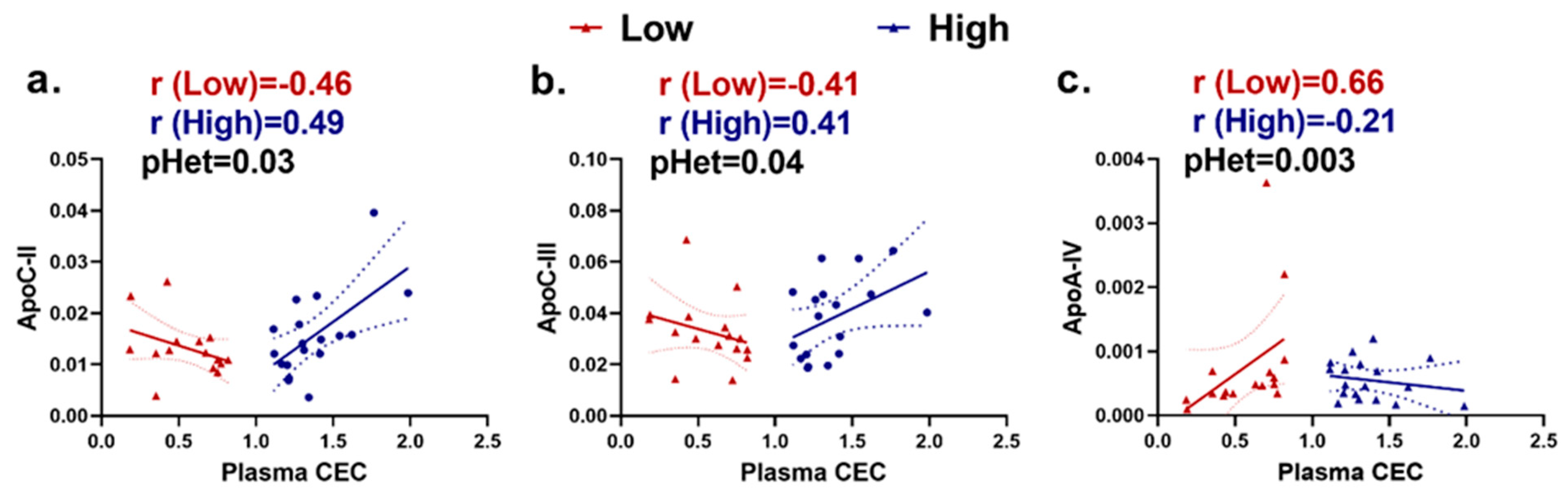
2.2. ApoA-I-Associated Proteins Are Heterogeneously Correlated with Plasma Cholesterol Efflux Capacity (CEC) in Extremely High versus Extremely Low CEC Groups
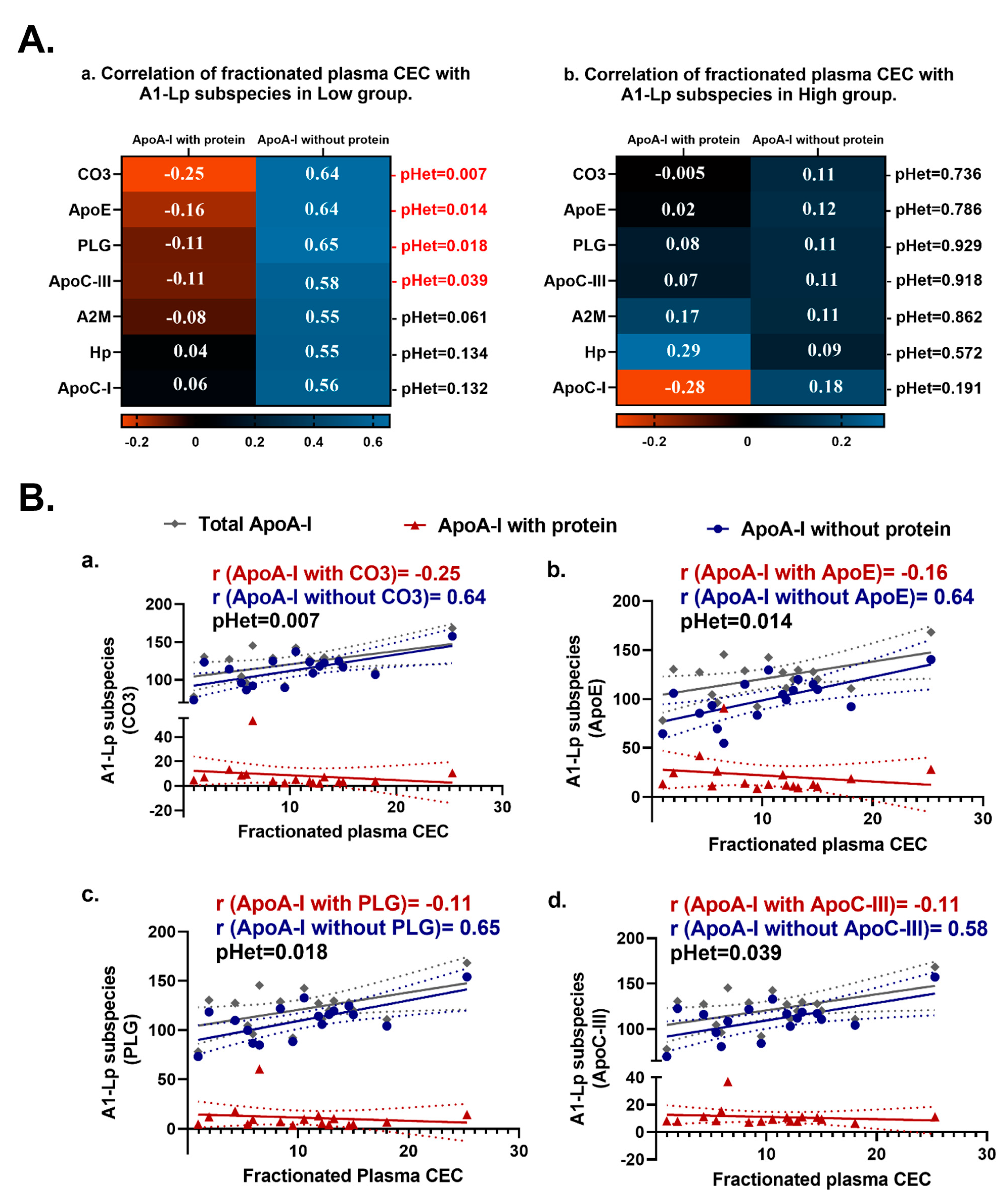
2.3. ApoA-I Containing Lipoprotein (AI-Lp) Subspecies with versus AI-Lp Subspecies without Certain Proteins Display Heterogeneous Correlations with Fractionated CEC in the Extremely Low CEC Group but Not in the Extremely High CEC Group
2.4. Relationships between apoA-I-Associated Proteins and apoA-I Containing Lipoprotein (AI-Lp) Subspecies Display Heterogeneity between the Extremely High and Extremely Low CEC Groups
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Blood Collection and Storage
4.3. Affinity Enrichment and Targeted Proteomics Analysis of ApoA-I-Associated Lipoproteins
4.4. Measurement of ApoA-I-Containing Subspecies with and without Selected Protein (AI-Lp Subspecies)
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics—2021 Update: A Report From the American Heart Association. Circulation 2021, 143, e254–e743. [Google Scholar] [CrossRef] [PubMed]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and Reverse Cholesterol Transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Rosenson, R.S.; Brewer, H.B., Jr.; Davidson, W.S.; Fayad, Z.A.; Fuster, V.; Goldstein, J.; Hellerstein, M.; Jiang, X.C.; Phillips, M.C.; Rader, D.J.; et al. Cholesterol efflux and atheroprotection: Advancing the concept of reverse cholesterol transport. Circulation 2012, 125, 1905–1919. [Google Scholar] [CrossRef] [PubMed]
- Pownall, H.J.; Rosales, C.; Gillard, B.K.; Gotto, A.M., Jr. High-density lipoproteins, reverse cholesterol transport and atherogenesis. Nature reviews. Cardiology 2021, 18, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Briel, M.; Ferreira-Gonzalez, I.; You, J.J.; Karanicolas, P.J.; Akl, E.A.; Wu, P.; Blechacz, B.; Bassler, D.; Wei, X.; Sharman, A.; et al. Association between change in high density lipoprotein cholesterol and cardiovascular disease morbidity and mortality: Systematic review and meta-regression analysis. BMJ 2009, 338, b92. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.A. The complexity of HDL. Biochim. Biophys. Acta 2010, 1801, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Peloso, G.M.; Orho-Melander, M.; Frikke-Schmidt, R.; Barbalic, M.; Jensen, M.K.; Hindy, G.; Holm, H.; Ding, E.L.; Johnson, T.; et al. Plasma HDL cholesterol and risk of myocardial infarction: A mendelian randomisation study. Lancet 2012, 380, 572–580. [Google Scholar] [CrossRef]
- Nicholls, S.J. CETP-Inhibition and HDL-Cholesterol: A Story of CV Risk or CV Benefit, or Both. Clin. Pharmacol. Ther. 2018, 104, 297–300. [Google Scholar] [CrossRef]
- Barter, P.J.; Caulfield, M.; Eriksson, M.; Grundy, S.M.; Kastelein, J.J.; Komajda, M.; Lopez-Sendon, J.; Mosca, L.; Tardif, J.C.; Waters, D.D.; et al. Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 2007, 357, 2109–2122. [Google Scholar] [CrossRef]
- Investigators, A.-H.; Boden, W.E.; Probstfield, J.L.; Anderson, T.; Chaitman, B.R.; Desvignes-Nickens, P.; Koprowicz, K.; McBride, R.; Teo, K.; Weintraub, W. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N. Engl. J. Med. 2011, 365, 2255–2267. [Google Scholar] [CrossRef]
- Schwartz, G.G.; Olsson, A.G.; Abt, M.; Ballantyne, C.M.; Barter, P.J.; Brumm, J.; Chaitman, B.R.; Holme, I.M.; Kallend, D.; Leiter, L.A.; et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med. 2012, 367, 2089–2099. [Google Scholar] [CrossRef]
- Group, H.T.C.; Landray, M.J.; Haynes, R.; Hopewell, J.C.; Parish, S.; Aung, T.; Tomson, J.; Wallendszus, K.; Craig, M.; Jiang, L.; et al. Effects of extended-release niacin with laropiprant in high-risk patients. N. Engl. J. Med. 2014, 371, 203–212. [Google Scholar] [CrossRef]
- Von Eckardstein, A.; Nordestgaard, B.G.; Remaley, A.T.; Catapano, A.L. High-density lipoprotein revisited: Biological functions and clinical relevance. Eur. Heart J. 2023, 44, 1394–1407. [Google Scholar] [CrossRef] [PubMed]
- Rye, K.A.; Barter, P.J. Cardioprotective functions of HDLs. J. Lipid Res. 2014, 55, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Cuchel, M.; de la Llera-Moya, M.; Rodrigues, A.; Burke, M.F.; Jafri, K.; French, B.C.; Phillips, J.A.; Mucksavage, M.L.; Wilensky, R.L.; et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N. Engl. J. Med. 2011, 364, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, Y.; Ding, D.; Li, X.; Yang, Y.; Li, Q.; Zheng, Y.; Wang, D.; Ling, W. Cholesterol efflux capacity is an independent predictor of all-cause and cardiovascular mortality in patients with coronary artery disease: A prospective cohort study. Atherosclerosis 2016, 249, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Saleheen, D.; Scott, R.; Javad, S.; Zhao, W.; Rodrigues, A.; Picataggi, A.; Lukmanova, D.; Mucksavage, M.L.; Luben, R.; Billheimer, J.; et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: A prospective case-control study. Lancet Diabetes Endocrinol. 2015, 3, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Demler, O.V.; Adelman, S.J.; Collins, H.L.; Glynn, R.J.; Ridker, P.M.; Rader, D.J.; Mora, S. Cholesterol Efflux Capacity, High-Density Lipoprotein Particle Number, and Incident Cardiovascular Events: An Analysis From the JUPITER Trial (Justification for the Use of Statins in Prevention: An Intervention Trial Evaluating Rosuvastatin). Circulation 2017, 135, 2494–2504. [Google Scholar] [CrossRef]
- Shea, S.; Stein, J.H.; Jorgensen, N.W.; McClelland, R.L.; Tascau, L.; Shrager, S.; Heinecke, J.W.; Yvan-Charvet, L.; Tall, A.R. Cholesterol Mass Efflux Capacity, Incident Cardiovascular Disease, and Progression of Carotid Plaque. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 89–96. [Google Scholar] [CrossRef]
- Ebtehaj, S.; Gruppen, E.G.; Bakker, S.J.L.; Dullaart, R.P.F.; Tietge, U.J.F. HDL (High-Density Lipoprotein) Cholesterol Efflux Capacity Is Associated with Incident Cardiovascular Disease in the General Population. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1874–1883. [Google Scholar] [CrossRef]
- Lee, J.J.; Chi, G.; Fitzgerald, C.; Kazmi, S.H.A.; Kalayci, A.; Korjian, S.; Duffy, D.; Shaunik, A.; Kingwell, B.; Yeh, R.W.; et al. Cholesterol Efflux Capacity and Its Association with Adverse Cardiovascular Events: A Systematic Review and Meta-Analysis. Front. Cardiovasc. Med. 2021, 8, 774418. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, A.; Khera, A.; Berry, J.D.; Givens, E.G.; Ayers, C.R.; Wedin, K.E.; Neeland, I.J.; Yuhanna, I.S.; Rader, D.R.; de Lemos, J.A.; et al. HDL cholesterol efflux capacity and incident cardiovascular events. N. Engl. J. Med. 2014, 371, 2383–2393. [Google Scholar] [CrossRef] [PubMed]
- Ronsein, G.E.; Vaisar, T. Inflammation, remodeling, and other factors affecting HDL cholesterol efflux. Curr. Opin. Lipidol. 2017, 28, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Vaisar, T.; Tang, C.; Babenko, I.; Hutchins, P.; Wimberger, J.; Suffredini, A.F.; Heinecke, J.W. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity. J. Lipid Res. 2015, 56, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Edelstein, C.; Pfaffinger, D.; Scanu, A.M. Advantages and limitations of density gradient ultracentrifugation in the fractionation of human serum lipoproteins: Role of salts and sucrose. J. Lipid Res. 1984, 25, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Kunitake, S.T.; Kane, J.P. Factors affecting the integrity of high density lipoproteins in the ultracentrifuge. J. Lipid Res. 1982, 23, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Collier, T.S.; Jin, Z.; Topbas, C.; Bystrom, C. Rapid Affinity Enrichment of Human Apolipoprotein A-I Associated Lipoproteins for Proteome Analysis. J. Proteome Res. 2018, 17, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Collier, T.S.; Jin, Z.; Lyass, A.; Li, Y.; Ibrahim, N.E.; Mukai, R.; McCarthy, C.P.; Massaro, J.M.; D’Agostino, R.B., Sr.; et al. Association of an HDL Apolipoproteomic Score with Coronary Atherosclerosis and Cardiovascular Death. J. Am. Coll. Cardiol. 2019, 73, 2135–2145. [Google Scholar] [CrossRef]
- Furtado, J.D.; Yamamoto, R.; Melchior, J.T.; Andraski, A.B.; Gamez-Guerrero, M.; Mulcahy, P.; He, Z.; Cai, T.; Davidson, W.S.; Sacks, F.M. Distinct Proteomic Signatures in 16 HDL (High-Density Lipoprotein) Subspecies. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2827–2842. [Google Scholar] [CrossRef]
- Sacks, F.M.; Liang, L.; Furtado, J.D.; Cai, T.; Davidson, W.S.; He, Z.; McClelland, R.L.; Rimm, E.B.; Jensen, M.K. Protein-Defined Subspecies of HDLs (High-Density Lipoproteins) and Differential Risk of Coronary Heart Disease in 4 Prospective Studies. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 2714–2727. [Google Scholar] [CrossRef]
- El-Ghazali, A.; Deodhar, S.; Saldanha, S.; Smyth, B.; Izbrand, M.; Gangwar, A.; Pahlavani, M.; Rohatgi, A. Molecular Patterns of Extreme and Persistent Cholesterol Efflux Capacity. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2588–2597. [Google Scholar] [CrossRef]
- Davidson, W.S.; Cooke, A.L.; Swertfeger, D.K.; Shah, A.S. The Difference Between High Density Lipoprotein Subfractions and Subspecies: An Evolving Model in Cardiovascular Disease and Diabetes. Curr. Atheroscler. Rep. 2021, 23, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Tang, W.H.; Mosior, M.K.; Huang, Y.; Wu, Y.; Matter, W.; Gao, V.; Schmitt, D.; Didonato, J.A.; Fisher, E.A.; et al. Paradoxical association of enhanced cholesterol efflux with increased incident cardiovascular risks. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1696–1705. [Google Scholar] [CrossRef] [PubMed]
- Amar, M.J.; D’Souza, W.; Turner, S.; Demosky, S.; Sviridov, D.; Stonik, J.; Luchoomun, J.; Voogt, J.; Hellerstein, M.; Sviridov, D.; et al. 5A apolipoprotein mimetic peptide promotes cholesterol efflux and reduces atherosclerosis in mice. J. Pharmacol. Exp. Ther. 2010, 334, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.W.; Tsai, M.C.; Chern, C.Y.; Tsao, T.P.; Lin, F.Y.; Chen, S.J.; Tsui, P.F.; Liu, Y.W.; Lu, H.J.; Wu, W.L.; et al. A chalcone derivative, 1m-6, exhibits atheroprotective effects by increasing cholesterol efflux and reducing inflammation-induced endothelial dysfunction. Br. J. Pharmacol. 2020, 177, 5375–5392. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Guo, Y.; Ruan, G.; Li, X. Metformin promotes cholesterol efflux in macrophages by up-regulating FGF21 expression: A novel anti-atherosclerotic mechanism. Lipids Health Dis. 2016, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Kingwell, B.A.; Nicholls, S.J.; Velkoska, E.; Didichenko, S.A.; Duffy, D.; Korjian, S.; Gibson, C.M. Antiatherosclerotic Effects of CSL112 Mediated by Enhanced Cholesterol Efflux Capacity. J. Am. Heart Assoc. 2022, 11, e024754. [Google Scholar] [CrossRef] [PubMed]
- Davidson, W.S.; Shah, A.S.; Sexmith, H.; Gordon, S.M. The HDL Proteome Watch: Compilation of studies leads to new insights on HDL function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2022, 1867, 159072. [Google Scholar] [CrossRef]
- Rhainds, D.; Tardif, J.C. From HDL-cholesterol to HDL-function: Cholesterol efflux capacity determinants. Curr. Opin. Lipidol. 2019, 30, 101–107. [Google Scholar] [CrossRef]
- Florea, G.; Tudorache, I.F.; Fuior, E.V.; Ionita, R.; Dumitrescu, M.; Fenyo, I.M.; Bivol, V.G.; Gafencu, A.V. Apolipoprotein A-II, a Player in Multiple Processes and Diseases. Biomedicines 2022, 10, 1578. [Google Scholar] [CrossRef]
- Cohen, R.D.; Castellani, L.W.; Qiao, J.H.; Van Lenten, B.J.; Lusis, A.J.; Reue, K. Reduced aortic lesions and elevated high density lipoprotein levels in transgenic mice overexpressing mouse apolipoprotein A-IV. J. Clin. Investig. 1997, 99, 1906–1916. [Google Scholar] [CrossRef]
- Remaley, A.T.; Stonik, J.A.; Demosky, S.J.; Neufeld, E.B.; Bocharov, A.V.; Vishnyakova, T.G.; Eggerman, T.L.; Patterson, A.P.; Duverger, N.J.; Santamarina-Fojo, S.; et al. Apolipoprotein specificity for lipid efflux by the human ABCAI transporter. Biochem. Biophys. Res. Commun. 2001, 280, 818–823. [Google Scholar] [CrossRef] [PubMed]
- Pearson, K.; Saito, H.; Woods, S.C.; Lund-Katz, S.; Tso, P.; Phillips, M.C.; Davidson, W.S. Structure of human apolipoprotein A-IV: A distinct domain architecture among exchangeable apolipoproteins with potential functional implications. Biochemistry 2004, 43, 10719–10729. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Liu, A.; Wang, S.; Wang, T.; Hu, D.; Wu, S.; Peng, D. ApoCIII enrichment in HDL impairs HDL-mediated cholesterol efflux capacity. Sci. Rep. 2017, 7, 2312. [Google Scholar] [CrossRef] [PubMed]
- The TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute; Crosby, J.; Peloso, G.M.; Auer, P.L.; Crosslin, D.R.; Stitziel, N.O.; Lange, L.A.; Lu, Y. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 2014, 371, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Liao, C.W.; Meng, S.W.; Wu, W.K.; Chiang, J.Y.; Wu, M.S. Lipids and Lipoproteins in Health and Disease: Focus on Targeting Atherosclerosis. Biomedicines 2021, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ronsein, G.E.; Tang, C.; Jarvik, G.P.; Davidson, W.S.; Kothari, V.; Song, H.D.; Segrest, J.P.; Bornfeldt, K.E.; Heinecke, J.W. Diabetes Impairs Cellular Cholesterol Efflux From ABCA1 to Small HDL Particles. Circ. Res. 2020, 127, 1198–1210. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gates, K.P.; Fang, L.; Amar, M.J.; Schneider, D.A.; Geng, H.; Huang, W.; Kim, J.; Pattison, J.; Zhang, J.; et al. Apoc2 loss-of-function zebrafish mutant as a genetic model of hyperlipidemia. Dis. Model. Mech. 2015, 8, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Low-Kam, C.; Rhainds, D.; Lo, K.S.; Barhdadi, A.; Boule, M.; Alem, S.; Pedneault-Gagnon, V.; Rheaume, E.; Dube, M.P.; Busseuil, D.; et al. Variants at the APOE/C1/C2/C4 Locus Modulate Cholesterol Efflux Capacity Independently of High-Density Lipoprotein Cholesterol. J. Am. Heart Assoc. 2018, 7, e009545. [Google Scholar] [CrossRef]
- Gordon, S.M.; Deng, J.; Tomann, A.B.; Shah, A.S.; Lu, L.J.; Davidson, W.S. Multi-dimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies. Mol. Cell Proteom. 2013, 12, 3123–3134. [Google Scholar] [CrossRef]
- Li, H.; Gordon, S.M.; Zhu, X.; Deng, J.; Swertfeger, D.K.; Davidson, W.S.; Lu, L.J. Network-Based Analysis on Orthogonal Separation of Human Plasma Uncovers Distinct High Density Lipoprotein Complexes. J. Proteome Res. 2015, 14, 3082–3094. [Google Scholar] [CrossRef] [PubMed]
- Rye, K.A.; Barter, P.J. Predictive value of different HDL particles for the protection against or risk of coronary heart disease. Biochim. Biophys. Acta 2012, 1821, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Aroner, S.A.; Mukamal, K.J.; Furtado, J.D.; Post, W.S.; Tsai, M.Y.; Tjonneland, A.; Polak, J.F.; Rimm, E.B.; Overvad, K.; et al. High-Density Lipoprotein Subspecies Defined by Presence of Apolipoprotein C-III and Incident Coronary Heart Disease in Four Cohorts. Circulation 2018, 137, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Alaupovic, P.; Moye, L.A.; Cole, T.G.; Sussex, B.; Stampfer, M.J.; Pfeffer, M.A.; Braunwald, E. VLDL, apolipoproteins B, CIII, and E, and risk of recurrent coronary events in the Cholesterol and Recurrent Events (CARE) trial. Circulation 2000, 102, 1886–1892. [Google Scholar] [CrossRef] [PubMed]
- Vaisar, T.; Pennathur, S.; Green, P.S.; Gharib, S.A.; Hoofnagle, A.N.; Cheung, M.C.; Byun, J.; Vuletic, S.; Kassim, S.; Singh, P.; et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 2007, 117, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Green, P.S.; Vaisar, T.; Pennathur, S.; Kulstad, J.J.; Moore, A.B.; Marcovina, S.; Brunzell, J.; Knopp, R.H.; Zhao, X.Q.; Heinecke, J.W. Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation 2008, 118, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Patel, N.; Abdelrahman, K.M.; Ling, C.; Reimund, M.; Graziano, G.; Sampson, M.; Playford, M.P.; Dey, A.K.; Reddy, A.; et al. Complex association of apolipoprotein E-containing HDL with coronary artery disease burden in cardiovascular disease. JCI Insight 2022, 7, e159577. [Google Scholar] [CrossRef] [PubMed]
- Pamir, N.; Hutchins, P.; Ronsein, G.; Vaisar, T.; Reardon, C.A.; Getz, G.S.; Lusis, A.J.; Heinecke, J.W. Proteomic analysis of HDL from inbred mouse strains implicates APOE associated with HDL in reduced cholesterol efflux capacity via the ABCA1 pathway. J. Lipid Res. 2016, 57, 246–257. [Google Scholar] [CrossRef]
- Lindstedt, L.; Kovanen, P.T. Plasmin and kallikrein reduce HDL-induced cholesterol efflux from foam cells. Biochem. Biophys. Res. Commun. 2000, 277, 552–557. [Google Scholar] [CrossRef]
- Pamir, N.; Hutchins, P.M.; Ronsein, G.E.; Wei, H.; Tang, C.; Das, R.; Vaisar, T.; Plow, E.; Schuster, V.; Koschinsky, M.L.; et al. Plasminogen promotes cholesterol efflux by the ABCA1 pathway. JCI Insight 2017, 2, e92176. [Google Scholar] [CrossRef]
- Gordon, S.M.; Chung, J.H.; Playford, M.P.; Dey, A.K.; Sviridov, D.; Seifuddin, F.; Chen, Y.C.; Pirooznia, M.; Chen, M.Y.; Mehta, N.N.; et al. High density lipoprotein proteome is associated with cardiovascular risk factors and atherosclerosis burden as evaluated by coronary CT angiography. Atherosclerosis 2018, 278, 278–285. [Google Scholar] [CrossRef]
- Victor, R.G.; Haley, R.W.; Willett, D.L.; Peshock, R.M.; Vaeth, P.C.; Leonard, D.; Basit, M.; Cooper, R.S.; Iannacchione, V.G.; Visscher, W.A.; et al. The Dallas Heart Study: A population-based probability sample for the multidisciplinary study of ethnic differences in cardiovascular health. Am. J. Cardiol. 2004, 93, 1473–1480. [Google Scholar] [CrossRef]


| Variable | Persistent High Efflux (N = 19) | Persistent Low Efflux (N = 17) | p-Value |
|---|---|---|---|
| Age, years | 56 (51–67) | 64 (58–71) | 0.12 |
| Female sex (%) | 68% (13) | 71% (12) | 0.89 |
| Hispanic ethnicity (%) | 21% (4) | 6% (1) | 0.24 |
| Black race (%) | 63% (12) | 59% (10) | 0.79 |
| Variable | Persistent High Efflux (N = 19) | Persistent Low Efflux (N = 17) | p-Value |
|---|---|---|---|
| Hypercholesterolemia (%) | 50% (9) | 53% (9) | 0.86 |
| Diabetes (%) | 33% (6) | 24% (4) | 0.52 |
| Heart disease (%) | 11% (2) | 0% (0) | 0.15 |
| Hypertension (%) | 44% (8) | 71% (12) | 0.12 |
| Menopausal (% of women) | 84% (11) | 92% (11) | 0.59 |
| Current smoker (%) | 21% (4) | 6% (1) | 0.19 |
| History of alcohol intake (%) | 68% (13) | 71% (12) | 0.89 |
| Blood pressure medication (%) | 47% (9) | 69% (11) | 0.20 |
| Glucose-lowering medication (%) | 26% (5) | 25% (4) | 0.93 |
| Lipid medication (%) | 42% (8) | 37% (6) | 0.78 |
| BMI | 30.4 (26.3–35.4) | 29.8 (26.0–34.6) | 0.96 |
| Total cholesterol, mg/dL | 178 (159–247) | 154 (146–197) | 0.27 |
| LDL-C, mg/dL | 102 (82–163) | 88 (81–126) | 0.21 |
| Non-HDL-C, mg/dL | 120 (103–198) | 107 (95–146) | 0.09 |
| Triglycerides, mg/dL | 120 (97–226) | 97 (75–117) | 0.04 |
| HDL-C, mg/dL | 48 (41–58) | 55 (49–61) | 0.27 |
| Apolipoprotein A-I, mg/dL | 154 (148–182) | 155 (149–171) | 0.92 |
| Glucose, mg/dL | 103 (89–125) | 95 (90–104) | 0.59 |
| Hemoglobin A1C, % | 5.7% (5.5–6.8) | 5.8% (5.5–5.9) | 0.61 |
| Creatinine, mg/dL | 0.88 (0.70–0.98) | 0.81 (0.58–0.92) | 0.33 |
| Total protein, g/dL | 7.3 (7.0–7.5) | 7.3 (6.9–7.3) | 0.38 |
| Albumin, g/dL | 4.4 (4.2–4.5) | 4.5 (4.1–4.6) | 0.34 |
| Hemoglobin, g/dL | 13.6 (12.9–14.9) | 13.7 (13.0–14.3) | 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gangwar, A.; Deodhar, S.S.; Saldanha, S.; Melander, O.; Abbasi, F.; Pearce, R.W.; Collier, T.S.; McPhaul, M.J.; Furtado, J.D.; Sacks, F.M.; et al. Proteomic Determinants of Variation in Cholesterol Efflux: Observations from the Dallas Heart Study. Int. J. Mol. Sci. 2023, 24, 15526. https://doi.org/10.3390/ijms242115526
Gangwar A, Deodhar SS, Saldanha S, Melander O, Abbasi F, Pearce RW, Collier TS, McPhaul MJ, Furtado JD, Sacks FM, et al. Proteomic Determinants of Variation in Cholesterol Efflux: Observations from the Dallas Heart Study. International Journal of Molecular Sciences. 2023; 24(21):15526. https://doi.org/10.3390/ijms242115526
Chicago/Turabian StyleGangwar, Anamika, Sneha S. Deodhar, Suzanne Saldanha, Olle Melander, Fahim Abbasi, Ryan W. Pearce, Timothy S. Collier, Michael J. McPhaul, Jeremy D. Furtado, Frank M. Sacks, and et al. 2023. "Proteomic Determinants of Variation in Cholesterol Efflux: Observations from the Dallas Heart Study" International Journal of Molecular Sciences 24, no. 21: 15526. https://doi.org/10.3390/ijms242115526







