Tumorigenicity of EGFR- and/or HER2-Positive Breast Cancers Is Mediated by Recruitment of Tumor-Associated Macrophages
Abstract
:1. Introduction
2. Results
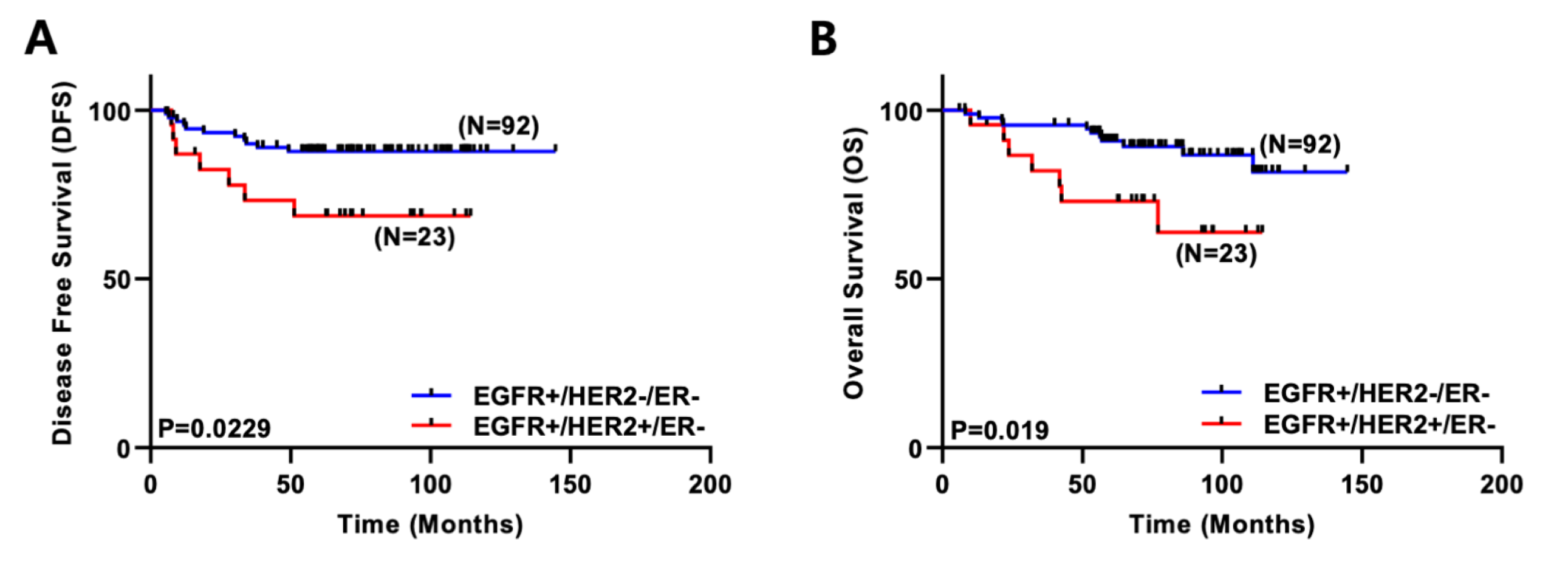
2.1. Co-Expression of EGFR and HER2 Correlates with Poor Survival in Breast Cancer Patients
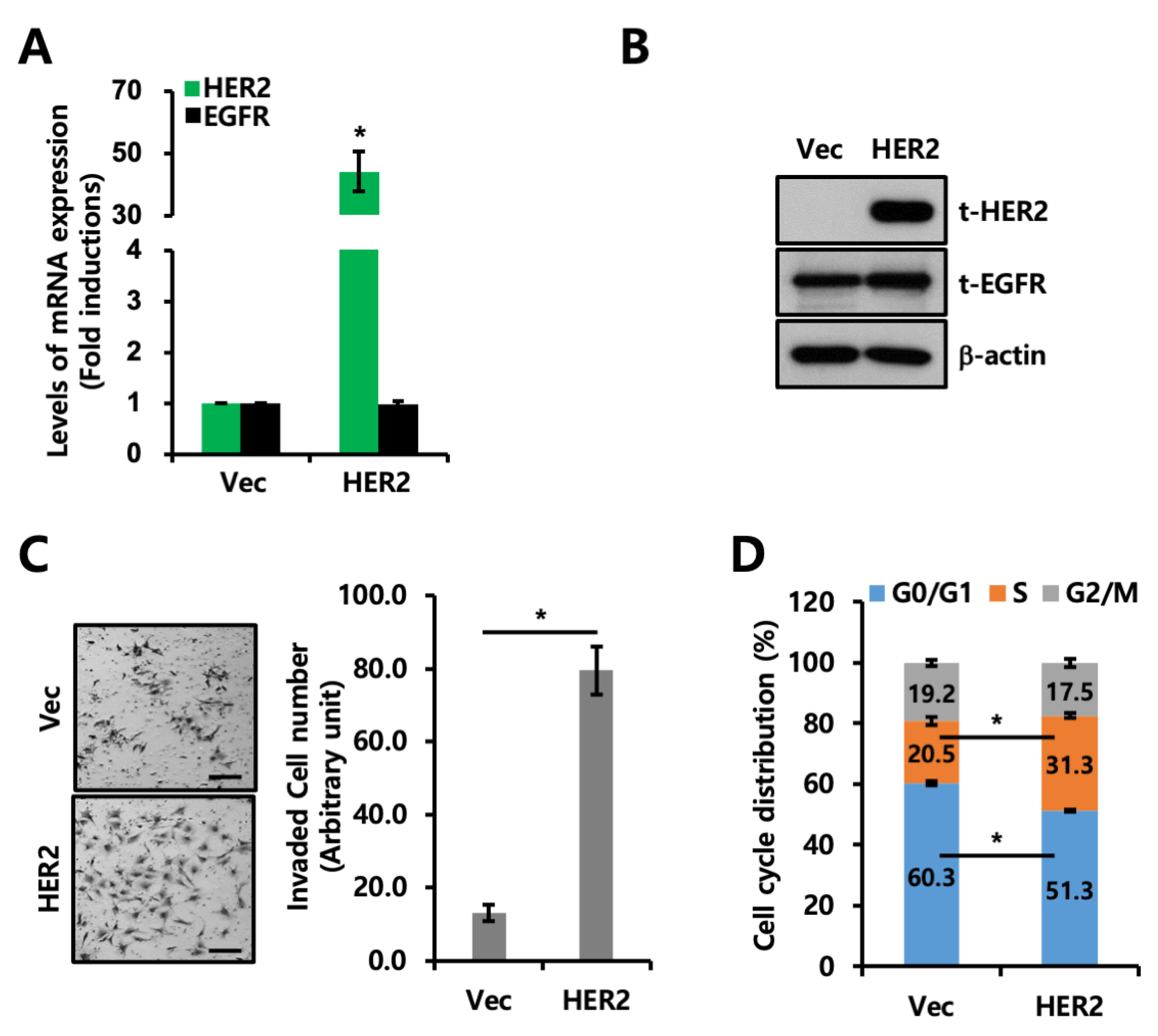
2.2. Co-Expression of EGFR and HER2 Promotes Invasion and Proliferation
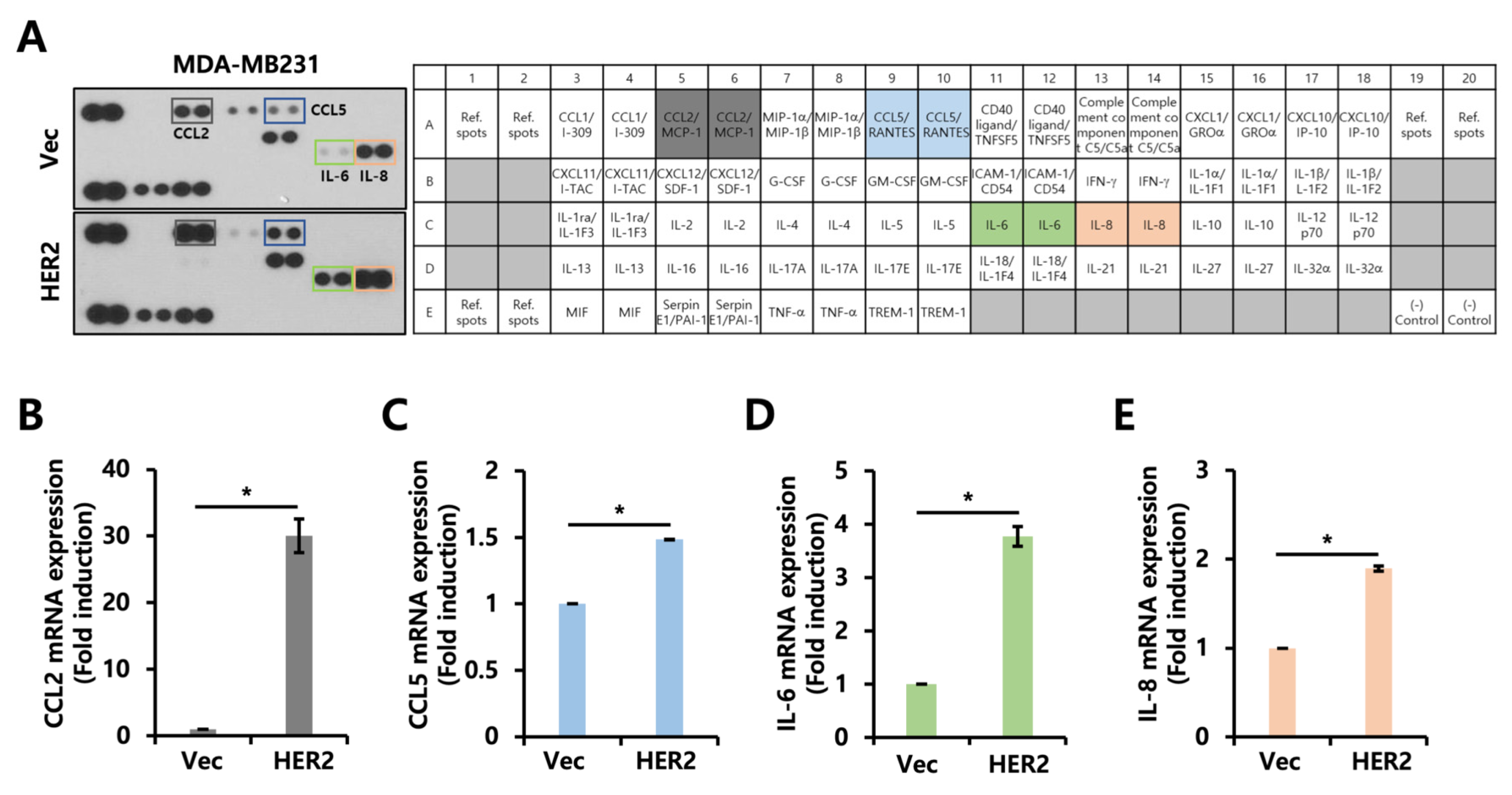
2.3. Cytokine Secretion Profile Is Altered by HER2 Overexpression
2.4. CCL2 Expression Is Downregulated by Neratinib, a Pan-HER Inhibitor
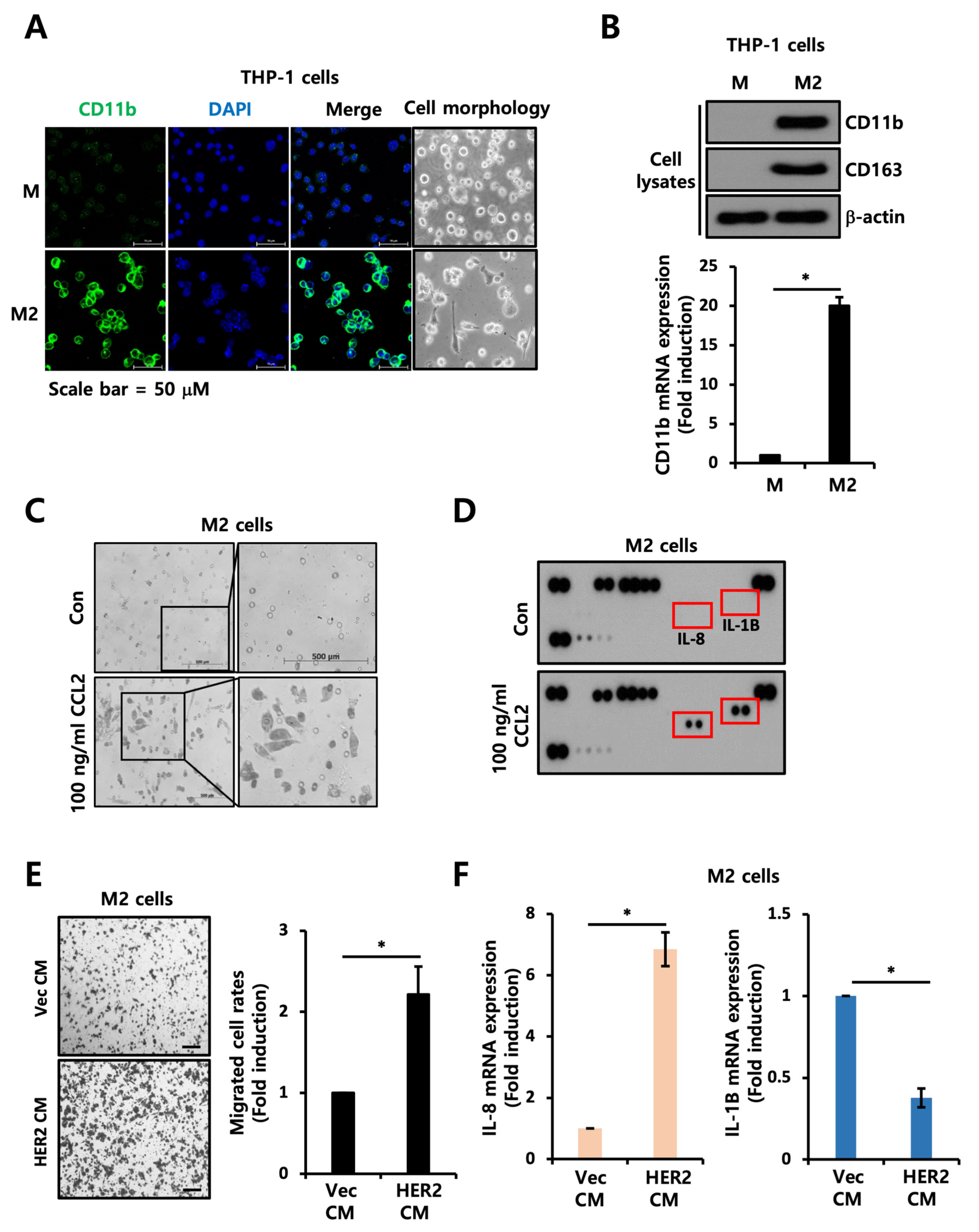
2.5. CCL2 Mediates Motility of TAMs
3. Discussion
4. Materials and Methods
4.1. Clinicopathological Characteristics of Breast Cancer Patients
4.2. Cell Culture
4.3. HER2 Knockdown Using siRNAs
4.4. Establishment of M2-Polarized THP-1 Macrophages
4.5. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
4.6. Western Blotting
4.7. Cell Invasion/Migration Assays
4.8. Cell Cycle Analysis
4.9. Human Cytokine Array
4.10. Enzyme-Linked Immunosorbent Assay (ELISA)
4.11. Immunofluorescence Staining
4.12. Immunohistochemistry (IHC)
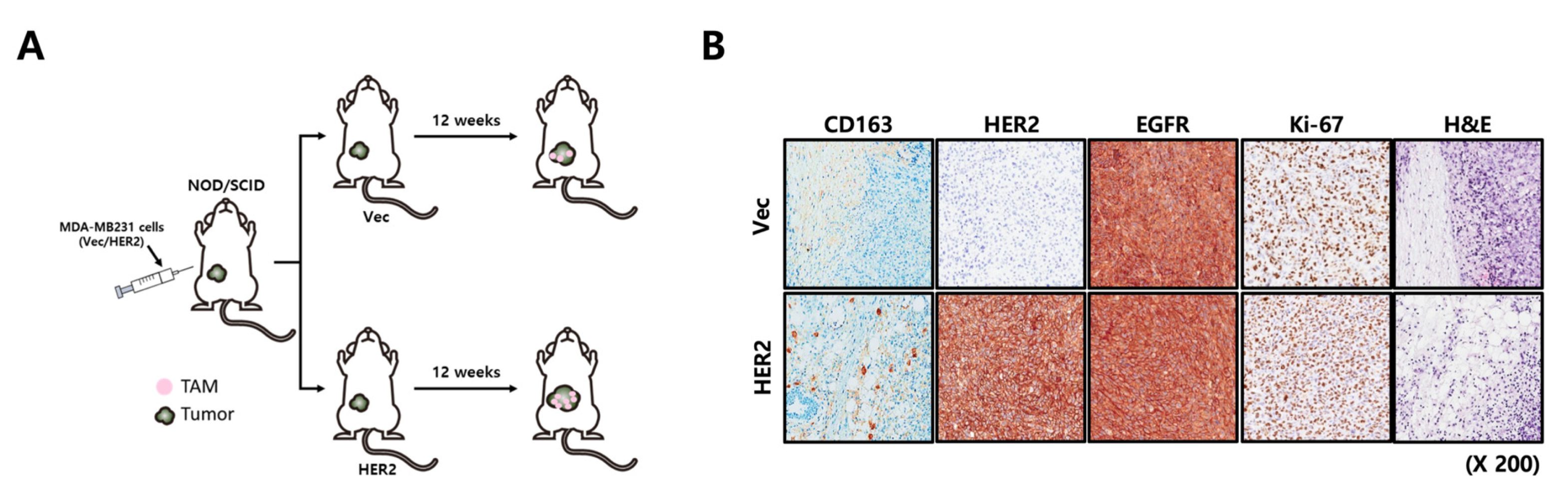
4.13. Xenograft Studies
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Fan, Q.; Zhang, Z.; Li, X.; Yu, H.; Meng, F. Basal-HER2 phenotype shows poorer survival than basal-like phenotype in hormone receptor-negative invasive breast cancers. Hum. Pathol. 2008, 39, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Bagaria, S.P.; Ray, P.S.; Wang, J.; Kropcho, L.; Chung, A.; Sim, M.S.; Shamonki, J.M.; Martino, S.; Cui, X.; Giuliano, A.E. Prognostic value of basal phenotype in HER2-overexpressing breast cancer. Ann. Surg. Oncol. 2012, 19, 935–940. [Google Scholar] [CrossRef]
- Weinberg, F.; Peckys, D.B.; de Jonge, N. EGFR Expression in HER2-Driven Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 9008. [Google Scholar] [CrossRef]
- Masuda, H.; Zhang, D.; Bartholomeusz, C.; Doihara, H.; Hortobagyi, G.N.; Ueno, N.T. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res. Treat. 2012, 136, 331–345. [Google Scholar] [CrossRef] [Green Version]
- Hsu, J.L.; Hung, M.C. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016, 35, 575–588. [Google Scholar] [CrossRef] [Green Version]
- Torregrosa, D.; Bolufer, P.; Lluch, A.; Lopez, J.A.; Barragan, E.; Ruiz, A.; Guillem, V.; Munarriz, B.; Garcia Conde, J. Prognostic significance of c-erbB-2/neu amplification and epidermal growth factor receptor (EGFR) in primary breast cancer and their relation to estradiol receptor (ER) status. Clin. Chim. Acta 1997, 262, 99–119. [Google Scholar] [CrossRef]
- Tsutsui, S.; Ohno, S.; Murakami, S.; Kataoka, A.; Kinoshita, J.; Hachitanda, Y. Prognostic value of the combination of epidermal growth factor receptor and c-erbB-2 in breast cancer. Surgery 2003, 133, 219–221. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Pollard, J.W. Macrophages define the invasive microenvironment in breast cancer. J. Leukoc. Biol. 2008, 84, 623–630. [Google Scholar] [CrossRef] [Green Version]
- Qiu, S.Q.; Waaijer, S.J.H.; Zwager, M.C.; de Vries, E.G.E.; van der Vegt, B.; Schroder, C.P. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat. Rev. 2018, 70, 178–189. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Dhillon, S. Neratinib in Early-Stage Breast Cancer: A Profile of Its Use in the EU. Clin. Drug Investig. 2019, 39, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Hao, Q.; Vadgama, J.V.; Wang, P. CCL2/CCR2 signaling in cancer pathogenesis. Cell Commun. Signal. 2020, 18, 82. [Google Scholar] [CrossRef]
- Roca, H.; Varsos, Z.S.; Sud, S.; Craig, M.J.; Ying, C.; Pienta, K.J. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J. Biol. Chem. 2009, 284, 34342–34354. [Google Scholar] [CrossRef] [Green Version]
- Tjiu, J.W.; Chen, J.S.; Shun, C.T.; Lin, S.J.; Liao, Y.H.; Chu, C.Y.; Tsai, T.F.; Chiu, H.C.; Dai, Y.S.; Inoue, H.; et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J. Investig. Dermatol. 2009, 129, 1016–1025. [Google Scholar] [CrossRef] [Green Version]
- Waugh, D.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Rebe, C.; Ghiringhelli, F. Interleukin-1beta and Cancer. Cancers 2020, 12, 1791. [Google Scholar] [CrossRef] [PubMed]
- Siziopikou, K.P.; Cobleigh, M. The basal subtype of breast carcinomas may represent the group of breast tumors that could benefit from EGFR-targeted therapies. Breast 2007, 16, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Livasy, C.A.; Karaca, G.; Nanda, R.; Tretiakova, M.S.; Olopade, O.I.; Moore, D.T.; Perou, C.M. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod. Pathol. 2006, 19, 264–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrelli, F.; Barni, S. Role of HER2-neu as a prognostic factor for survival and relapse in pT1a-bN0M0 breast cancer: A systematic review of the literature with a pooled-analysis. Med. Oncol. 2012, 29, 2586–2593. [Google Scholar] [CrossRef] [PubMed]
- Laakso, M.; Tanner, M.; Nilsson, J.; Wiklund, T.; Erikstein, B.; Kellokumpu-Lehtinen, P.; Malmstrom, P.; Wilking, N.; Bergh, J.; Isola, J. Basoluminal carcinoma: A new biologically and prognostically distinct entity between basal and luminal breast cancer. Clin. Cancer Res. 2006, 12, 4185–4191. [Google Scholar] [CrossRef] [Green Version]
- Guo, P.; Pu, T.; Chen, S.; Qiu, Y.; Zhong, X.; Zheng, H.; Chen, L.; Bu, H.; Ye, F. Breast cancers with EGFR and HER2 co-amplification favor distant metastasis and poor clinical outcome. Oncol. Lett. 2017, 14, 6562–6570. [Google Scholar] [CrossRef] [Green Version]
- Suo, Z.; Risberg, B.; Kalsson, M.G.; Willman, K.; Tierens, A.; Skovlund, E.; Nesland, J.M. EGFR family expression in breast carcinomas. c-erbB-2 and c-erbB-4 receptors have different effects on survival. J. Pathol. 2002, 196, 17–25. [Google Scholar] [CrossRef]
- Olayioye, M.A.; Neve, R.M.; Lane, H.A.; Hynes, N.E. The ErbB signaling network: Receptor heterodimerization in development and cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef] [Green Version]
- Mukohara, T. Mechanisms of resistance to anti-human epidermal growth factor receptor 2 agents in breast cancer. Cancer Sci. 2011, 102, 1–8. [Google Scholar] [CrossRef]
- Jeon, M.; Lee, J.; Nam, S.J.; Shin, I.; Lee, J.E.; Kim, S. Induction of fibronectin by HER2 overexpression triggers adhesion and invasion of breast cancer cells. Exp. Cell Res. 2015, 333, 116–126. [Google Scholar] [CrossRef]
- Jin, J.; Lin, J.; Xu, A.; Lou, J.; Qian, C.; Li, X.; Wang, Y.; Yu, W.; Tao, H. CCL2: An Important Mediator Between Tumor Cells and Host Cells in Tumor Microenvironment. Front. Oncol. 2021, 11, 722916. [Google Scholar] [CrossRef]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Kersten, K.; Coffelt, S.B.; Hoogstraat, M.; Verstegen, N.J.M.; Vrijland, K.; Ciampricotti, M.; Doornebal, C.W.; Hau, C.S.; Wellenstein, M.D.; Salvagno, C.; et al. Mammary tumor-derived CCL2 enhances pro-metastatic systemic inflammation through upregulation of IL1beta in tumor-associated macrophages. Oncoimmunology 2017, 6, e1334744. [Google Scholar] [CrossRef]
- Mahmoud, S.M.; Lee, A.H.; Paish, E.C.; Macmillan, R.D.; Ellis, I.O.; Green, A.R. Tumour-infiltrating macrophages and clinical outcome in breast cancer. J. Clin. Pathol. 2012, 65, 159–163. [Google Scholar] [CrossRef]
- Ali, H.R.; Chlon, L.; Pharoah, P.D.; Markowetz, F.; Caldas, C. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med. 2016, 13, e1002194. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Liu, M.; Du, X.; Li, S.; Li, H.; Li, X.; Li, Y.; Wang, Y.; Qin, Z.; Fu, Y.X.; et al. Intratumoral Delivery of IL-21 Overcomes Anti-Her2/Neu Resistance through Shifting Tumor-Associated Macrophages from M2 to M1 Phenotype. J. Immunol. 2015, 194, 4997–5006. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, Y.; Nelson, D.; Tian, S.; Mulvey, E.; Patel, B.; Conti, I.; Jaen, J.; Rollins, B.J. CCL2/CCR2 Regulates the Tumor Microenvironment in HER-2/neu-Driven Mammary Carcinomas in Mice. PLoS ONE 2016, 11, e0165595. [Google Scholar] [CrossRef] [Green Version]
- Linde, N.; Casanova-Acebes, M.; Sosa, M.S.; Mortha, A.; Rahman, A.; Farias, E.; Harper, K.; Tardio, E.; Reyes Torres, I.; Jones, J.; et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Vu, T.; Claret, F.X. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2012, 2, 62. [Google Scholar] [CrossRef] [Green Version]
- Harbeck, N.; Beckmann, M.W.; Rody, A.; Schneeweiss, A.; Muller, V.; Fehm, T.; Marschner, N.; Gluz, O.; Schrader, I.; Heinrich, G.; et al. HER2 Dimerization Inhibitor Pertuzumab—Mode of Action and Clinical Data in Breast Cancer. Breast Care 2013, 8, 49–55. [Google Scholar] [CrossRef]
- Choi, Y.L.; Oh, E.; Park, S.; Kim, Y.; Park, Y.H.; Song, K.; Cho, E.Y.; Hong, Y.C.; Choi, J.S.; Lee, J.E.; et al. Triple-negative, basal-like, and quintuple-negative breast cancers: Better prediction model for survival. BMC Cancer 2010, 10, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Variables | No. of Patients (%) |
|---|---|
| Age (years) | |
| <50 | 597 (62.8%) |
| ≥50 | 363 (37.2%) |
| Menopause | |
| Pre. | 588 (61.9%) |
| Post. | 362 (38.1%) |
| ER | |
| Negative (−) | 313 (35.9%) |
| Positive (+) | 558 (64.1%) |
| PR | |
| Negative (−) | 468 (53.1%) |
| Positive (+) | 413 (46.9%) |
| EGFR | |
| Negative (−) | 819 (86.3%) |
| Positive (+) | 130 (13.7%) |
| HER2 | |
| Negative (−) | 715 (75.3%) |
| Positive (+) | 235 (24.7%) |
| TP53 | |
| Negative (−) | 328 (41.8%) |
| Positive (+) | 457 (58.2%) |
| Operation | |
| BCS 1 | 359 (37.8%) |
| MRM 2 | 591 (62.2%) |
| T stage | |
| T1 | 391 (41.2%) |
| T2 | 494 (52.0%) |
| T3 | 62 (6.5%) |
| T4 | 3 (0.3%) |
| n stage | |
| N0 | 498 (52.4%) |
| N1 | 248 (26.1%) |
| N2 | 116 (12.2%) |
| N3 | 88 (9.3%) |
| Stage | |
| I | 258 (27.4%) |
| IIA | 332 (35.2%) |
| IIB | 135 (14.3%) |
| IIIA | 130 (13.8%) |
| IIIB | 2 (0.2%) |
| IIIC | 85 (9.0%) |
| Nuclear grade | |
| Low | 86 (9.1%) |
| Intermediate | 471 (49.6%) |
| High | 355 (37.4%) |
| Unknown | 38 (4.0%) |
| CK 5/6 | |
| Negative (−) | 819 (86.2%) |
| Positive (+) | 131 (13.8%) |
| Radiotherapy | |
| Yes | 532 (61.6%) |
| No | 332 (38.4%) |
| Unknown | 15 (1.7%) |
| Chemotherapy | |
| Yes | 794 (85.3%) |
| No | 137 (14.7%) |
| Unknown | 19 (2.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
You, D.; Kim, H.; Jeong, Y.; Yoon, S.Y.; Lo, E.; Kim, S.; Lee, J.E. Tumorigenicity of EGFR- and/or HER2-Positive Breast Cancers Is Mediated by Recruitment of Tumor-Associated Macrophages. Int. J. Mol. Sci. 2023, 24, 1443. https://doi.org/10.3390/ijms24021443
You D, Kim H, Jeong Y, Yoon SY, Lo E, Kim S, Lee JE. Tumorigenicity of EGFR- and/or HER2-Positive Breast Cancers Is Mediated by Recruitment of Tumor-Associated Macrophages. International Journal of Molecular Sciences. 2023; 24(2):1443. https://doi.org/10.3390/ijms24021443
Chicago/Turabian StyleYou, Daeun, Hyungjoo Kim, Yisun Jeong, Sun Young Yoon, Eunji Lo, Sangmin Kim, and Jeong Eon Lee. 2023. "Tumorigenicity of EGFR- and/or HER2-Positive Breast Cancers Is Mediated by Recruitment of Tumor-Associated Macrophages" International Journal of Molecular Sciences 24, no. 2: 1443. https://doi.org/10.3390/ijms24021443





