A New Face of the Old Gene: Deletion of the PssA, Encoding Monotopic Inner Membrane Phosphoglycosyl Transferase in Rhizobium leguminosarum, Leads to Diverse Phenotypes That Could Be Attributable to Downstream Effects of the Lack of Exopolysaccharide
Abstract
:1. Introduction
2. Results and Discussion
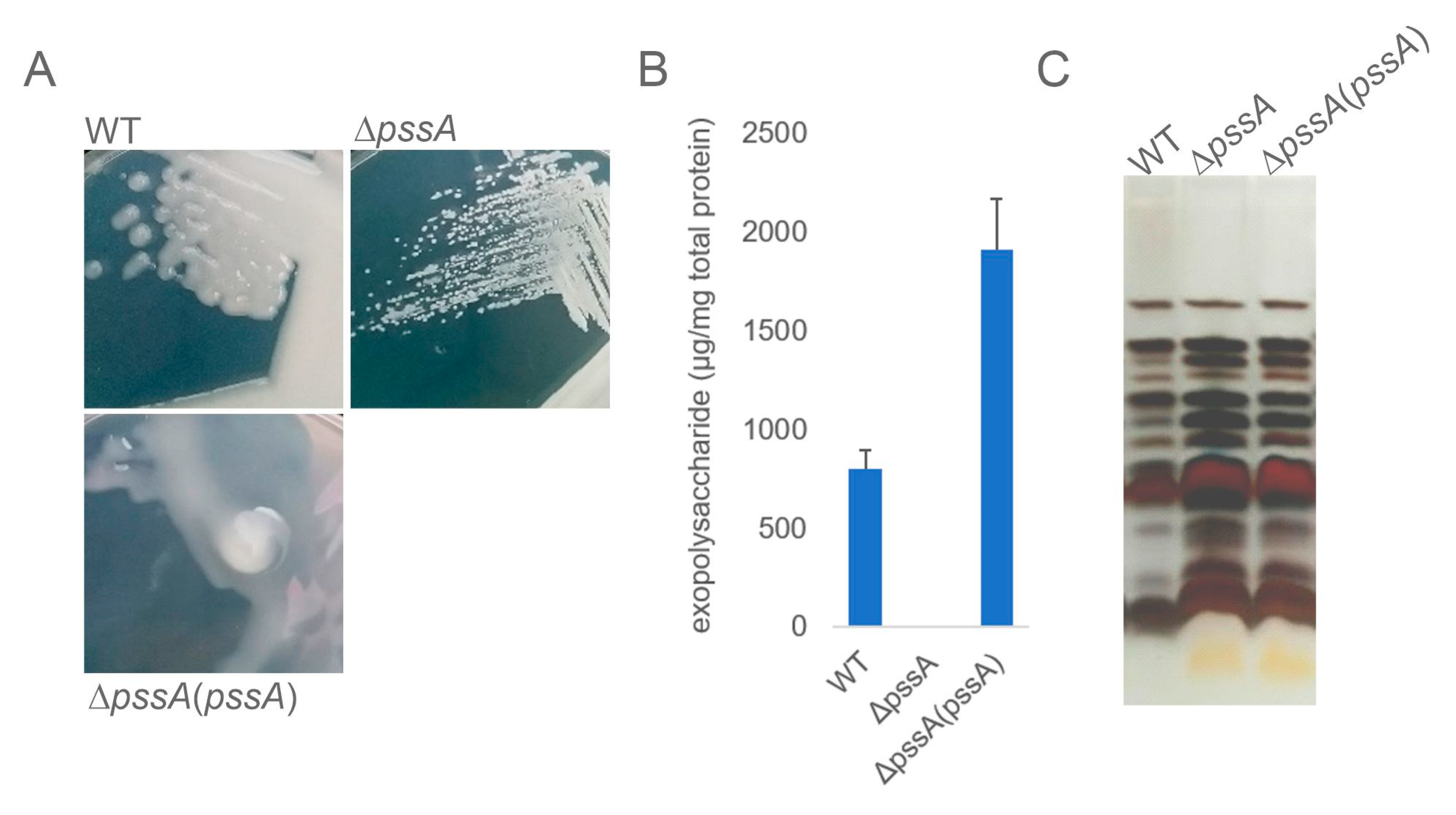
2.1. Deletion of PssA Abolishes EPS Biosynthesis and Affects Other Phenotypes of Rhizobium
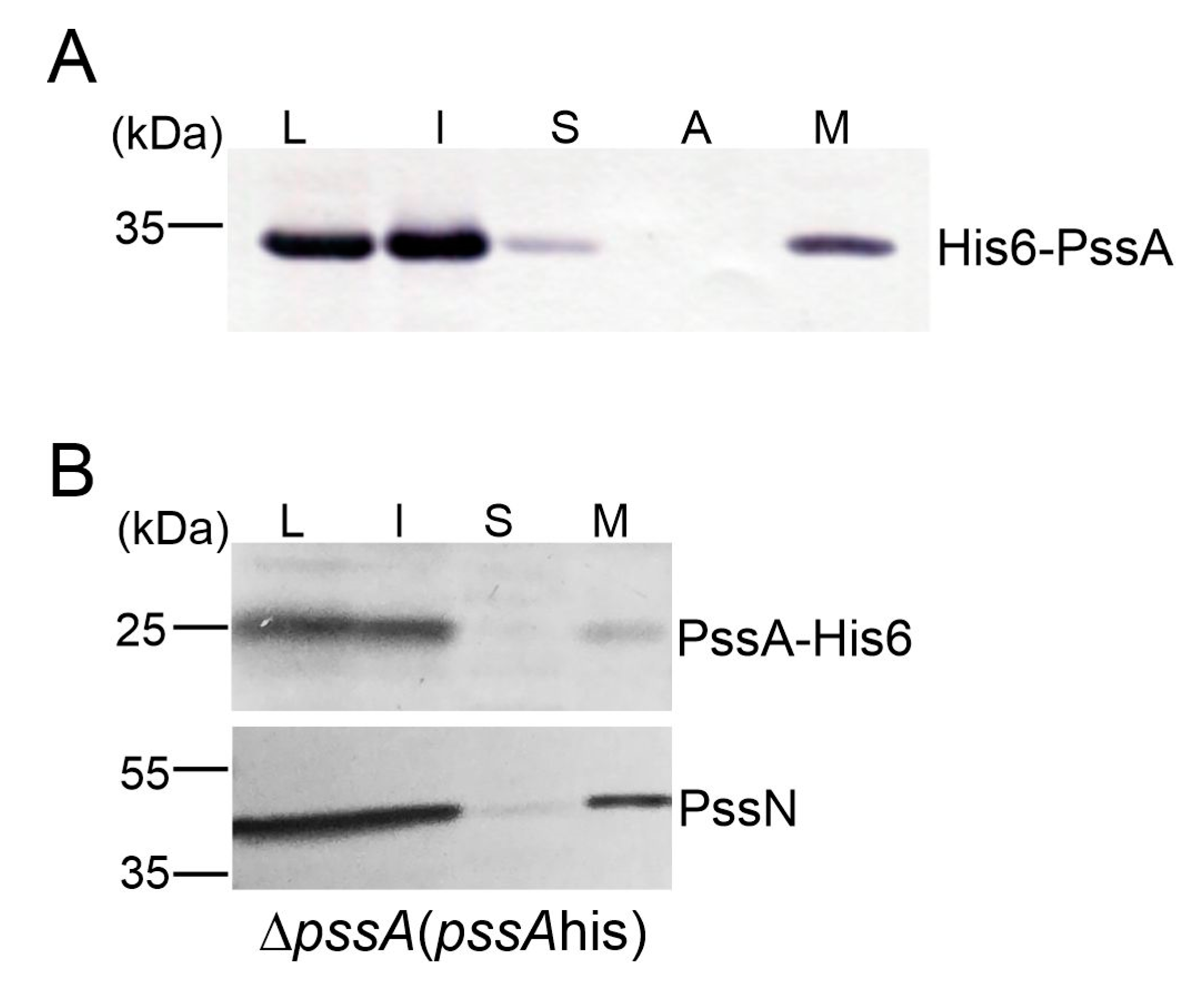
2.2. PssA Is an Inner Membrane Monotopic Protein with a Reentrant Helix-Break-Helix Motif
2.3. PssA Is a Phosphoglycosyltransferase Specific for UDP-Glucose
2.4. PssA Weakly Interacts with GTs Involved in the EPS Octasaccharide Synthesis
2.5. The Transcriptome of the ΔpssA Mutant Reveals Changes in the Expression of Genes Not Directly Related to EPS Biosynthesis
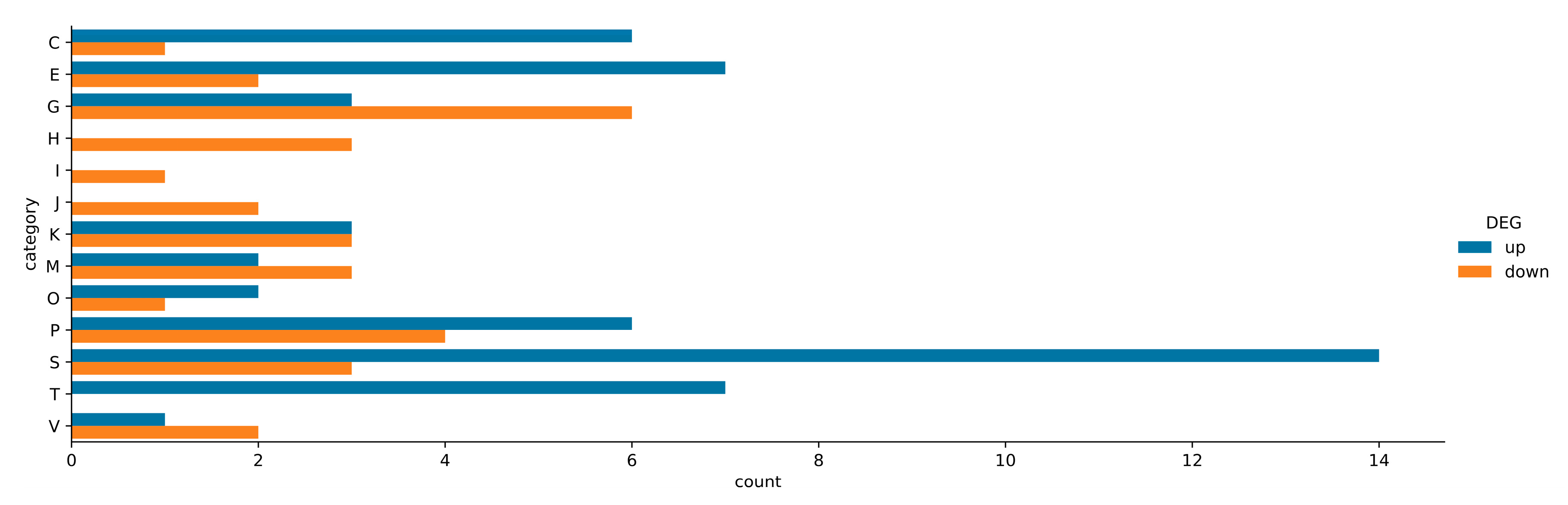
2.6. Assignment of ΔpssA Mutant DEGs to COG Categories and KEGG Pathways
2.7. Numerous Upregulated Genes of the ΔpssA Mutant Are Related to Amino Acid and Carbohydrate Transport and Metabolism as Well as Energy Conversion (COGs E, G, and C)
2.8. Activation of the Signal Transduction Mechanism (COG T) Comprises an Important Element of the ΔpssA Mutant Transcriptomic Response
2.9. Diverse Types of Genes Related to Stress Response Are Activated in the ΔpssA Mutant
2.10. Downregulated Genes of the ΔpssA Mutant Are Linked to the Transport and Metabolism of Coenzymes and Lipids as Well as Translation (COG H, I, J)
3. Materials and Methods
3.1. Bacterial Strains and Standard Culture Conditions
3.2. Bioinformatic Analyses
3.3. DNA Techniques
3.4. Construction of the RtTA1 ΔpssA Mutant and PssA Complemented Strains
3.5. Phenotypic Analyses
3.6. Construction of PhoA-LacZ Translational Fusions
3.7. Reporter Enzymes Quantitation
3.8. Overexpression and Purification of Recombinant PssA Protein
3.9. Protein Localization Study
3.10. UDP-Sugar Hydrolysis
3.11. Bacterial Two-Hybrid System
3.12. RNA Extraction
3.13. RNA Sequencing and Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donot, F.; Fontana, A.; Baccou, J.C.; Schorr-Galindo, S. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohyd. Polym. 2012, 87, 951–962. [Google Scholar] [CrossRef]
- Ghosh, P.K.; Maiti, T.K. Structure of extracellular polysaccharides (EPS) produced by rhizobia and their functions in legume–bacteria symbiosis. Achiev. Life Sci 2016, 10, 136–143. [Google Scholar] [CrossRef] [Green Version]
- Heredia-Ponce, Z.; de Vicente, A.; Cazorla, F.M.; Gutiérrez-Barranquero, J.A. Beyond the Wall: Exopolysaccharides in the Biofilm Lifestyle of Pathogenic and Beneficial Plant-Associated Pseudomonas. Microorganisms 2021, 9, 445. [Google Scholar] [CrossRef]
- Moscovici, M. Present and future medical applications of microbial exopolysaccharides. Front. Microbiol. 2015, 6, 1012. [Google Scholar] [CrossRef] [Green Version]
- Rehm, B.H. Bacterial polymers: Biosynthesis, modifications and applications. Nat. Rev. Microbiol. 2010, 8, 578–592. [Google Scholar] [CrossRef]
- Zayed, A.; Mansour, M.K.; Sedeek, M.S.; Habib, M.H.; Ulber, R.; Farag, M.A. Rediscovering bacterial exopolysaccharides of terrestrial and marine origins: Novel insights on their distribution, biosynthesis, biotechnological production, and future perspectives. Crit. Rev. Biotechnol. 2022, 42, 597–617. [Google Scholar] [CrossRef]
- Marczak, M.; Mazur, A.; Koper, P.; Żebracki, K.; Skorupska, A. Synthesis of rhizobial exopolysaccharides and their importance for symbiosis with legume plants. Genes 2017, 8, 360. [Google Scholar] [CrossRef] [Green Version]
- Kawaharada, Y.; Kelly, S.; Nielsen, M.W.; Hjuler, C.T.; Gysel, K.; Muszyński, A.; Carlson, R.W.; Thygesen, M.B.; Sandal, N.; Asmussen, M.H.; et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015, 523, 308–312. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Zhang, J. Bacterial exopolysaccharides: Chemical structures, gene clusters and genetic engineering. Int. J. Biol. Macromol. 2021, 173, 481–490. [Google Scholar] [CrossRef]
- Breedveld, M.W.; Cremers, H.C.; Batley, M.; Posthumus, M.A.; Zevenhuizen, L.P.; Wijffelman, C.A.; Zehnder, A.J. Polysaccharide synthesis in relation to nodulation behavior of Rhizobium leguminosarum. J. Bacteriol. 1993, 175, 750–757. [Google Scholar] [CrossRef]
- Król, J.E.; Mazur, A.; Marczak, M.; Skorupska, A. Syntenic arrangements of the surface polysaccharide biosynthesis genes in Rhizobium leguminosarum. Genomics 2007, 89, 237–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, M.A.; Darvill, A.G.; Albersheim, P. The degree of esterification and points of substitution by O-acetyl and O-(3-hydroxybutanoyl) groups in the acidic extracellular polysaccharides secreted by Rhizobium leguminosarum biovars viciae, trifolii, and phaseoli are not related to host range. J. Biol. Chem. 1991, 266, 9549–9555. [Google Scholar] [CrossRef] [PubMed]
- Philip-Hollingsworth, S.; Hollingsworth, R.I.; Dazzo, F.B. Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum. J. Biol. Chem. 1989, 264, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Robertsen, B.K.; Åman, P.; Darvill, A.G.; McNeil, M.; Albersheim, P. Host-symbiont interactions. V. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981, 67, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Pollock, T.J.; van Workum, W.A.; Thorne, L.; Mikolajczak, M.J.; Yamazaki, M.; Kijne, J.W.; Armentrout, R.W. Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J. Bacteriol. 1998, 180, 586–593. [Google Scholar] [CrossRef] [Green Version]
- Lukose, V.; Walvoort, M.T.C.; Imperiali, B. Bacterial phosphoglycosyl transferases: Initiators of glycan biosynthesis at the membrane interface. Glycobiology 2017, 27, 820–833. [Google Scholar] [CrossRef] [Green Version]
- Bossio, J.C.; Semino, C.E.; Iñón de Iannino, N.; Dankert, M.A. The in vitro biosynthesis of the exopolysaccharide produced by Rhizobium leguminosarum bv. trifolii, strain NA 30. Cell. Mol. Biol. 1996, 42, 737–758. [Google Scholar]
- Ivashina, T.V.; Ksenzenko, V.N. Exopolysaccharide biosynthesis in Rhizobium leguminosarum: From genes to functions. In The Complex World of Polysaccharides; Karunaratne, D.N., Ed.; IntechOpen: Rijeka, Croatia, 2012; pp. 99–126. [Google Scholar]
- Ivashina, T.V.; Khmelnitsky, M.I.; Shlyapnikov, M.G.; Kanapin, A.A.; Ksenzenko, V.N. The pss4 gene from Rhizobium leguminosarum bv viciae VF39: Cloning, sequence and the possible role in polysaccharide production and nodule formation. Gene 1994, 150, 111–116. [Google Scholar] [CrossRef]
- van Workum, W.A.; Canter Cremers, H.C.; Wijfjes, A.H.; van der Kolk, C.; Wijffelman, C.A.; Kijne, J.W. Cloning and characterization of four genes of Rhizobium leguminosarum bv. trifolii involved in exopolysaccharide production and nodulation. Mol. Plant Microbe Interact. 1997, 10, 290–301. [Google Scholar] [CrossRef]
- Lehrer, J.; Vigeant, K.A.; Tatar, L.D.; Valvano, M.A. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 2007, 189, 2618–2628. [Google Scholar] [CrossRef] [Green Version]
- Glover, K.J.; Weerapana, E.; Chen, M.M.; Imperiali, B. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 2006, 45, 5343–5350. [Google Scholar] [CrossRef]
- Entova, S.; Billod, J.M.; Swiecicki, J.M.; Martín-Santamaría, S.; Imperiali, B. Insights into the key determinants of membrane protein topology enable the identification of new monotopic folds. eLife 2018, 7, e40889. [Google Scholar] [CrossRef]
- Ksenzenko, V.N.; Ivashina, T.V.; Sadykhov, M.R.; Chatuev, B.M.; Khmelnitsky, M.I.; Kanapin, A.A.; Shlyapnikov, M.G. A role of the Rhizobium leguminosarum bv. viciae pss1 and pss4 genes in nitrogen fixing symbiosis. In First European Nitrogen Fixation Conference and Workshop on Safe Application of Genetically Modified Microorganisms in the Environment; Kiss, G.B., Endre, G., Eds.; Officina Press: Szeged, Hungary, 1994; pp. 142–146. [Google Scholar]
- Janczarek, M. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 2011, 12, 7898–7933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janczarek, M.; Urbanik-Sypniewska, T. Expression of the Rhizobium leguminosarum bv. trifolii pssA gene, involved in exopolysaccharide synthesis, is regulated by RosR, phosphate, and the carbon source. J. Bacteriol. 2013, 195, 3412–3423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerreiro, N.; Ksenzenko, V.N.; Djordjevic, M.A.; Ivashina, T.V.; Rolfe, B.G. Elevated levels of synthesis of over 20 proteins results after mutation of the Rhizobium leguminosarum exopolysaccharide synthesis gene pssA. J. Bacteriol. 2000, 182, 4521–4532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janczarek, M.; Jaroszuk-Ściseł, J.; Skorupska, A. Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie Van Leeuwenhoek 2009, 96, 471–486. [Google Scholar] [CrossRef]
- Janczarek, M.; Rachwał, K.; Turska-Szewczuk, A. A mutation in pssE affects exopolysaccharide synthesis by Rhizobium leguminosarum bv. trifolii, its surface properties, and symbiosis with clover. Plant Soil 2017, 417, 331–347. [Google Scholar] [CrossRef] [Green Version]
- Wielbo, J.; Mazur, A.; Król, J.E.; Marczak, M.; Skorupska, A. Environmental modulation of the pssTNOP gene expression in Rhizobium leguminosarum bv. trifolii. Can. J. Microbiol. 2004, 50, 201–211. [Google Scholar] [CrossRef]
- Janczarek, M.; Rachwał, K. Mutation in the pssA gene involved in exopolysaccharide synthesis leads to several physiological and symbiotic defects in Rhizobium leguminosarum bv. trifolii. Int. J. Mol. Sci. 2013, 14, 23711–23735. [Google Scholar] [CrossRef] [Green Version]
- Marx, C.J.; Lidstrom, M.E. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 2002, 33, 1062–1067. [Google Scholar] [CrossRef] [Green Version]
- Marczak, M.; Wójcik, M.; Żebracki, K.; Turska-Szewczuk, A.; Talarek, K.; Nowak, D.; Wawiórka, L.; Sieńczyk, M.; Łupicka-Słowik, A.; Bobrek, K.; et al. PssJ Is a Terminal Galactosyltransferase Involved in the Assembly of the Exopolysaccharide Subunit in Rhizobium Leguminosarum bv. Trifolii. Int. J. Mol. Sci. 2020, 21, 7764. [Google Scholar] [CrossRef] [PubMed]
- Marczak, M.; Żebracki, K.; Koper, P.; Turska-Szewczuk, A.; Mazur, A.; Wydrych, J.; Wójcik, M.; Skorupska, A. Mgl2 Is a hypothetical methyltransferase involved in exopolysaccharide production, biofilm formation, and motility in Rhizobium leguminosarum bv. trifolii. Mol. Plant Microbe Interact. 2019, 32, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Mazur, A.; Król, J.E.; Marczak, M.; Skorupska, A. Membrane topology of PssT, the transmembrane protein component of the type I exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii strain TA1. J. Bacteriol. 2003, 185, 2503–2511. [Google Scholar] [CrossRef] [Green Version]
- Islam, S.T.; Taylor, V.L.; Qi, M.; Lam, J.S. Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. mBio 2010, 1, e00189-10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ray, L.C.; Das, D.; Entova, S.; Lukose, V.; Lynch, A.J.; Imperiali, B.; Allen, K.N. Membrane association of monotopic phosphoglycosyl transferase underpins function. Nat. Chem. Biol. 2018, 14, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Ksenzenko, V.N.; Ivashina, T.V.; Dubeikovskaya, Z.A.; Ivanov, S.G.; Nanazashvili, M.B.; Druzhinina, T.N.; Kalinchuk, N.A.; Shibaev, V.N. The pssA gene encodes UDP-glucose: Polyprenyl phosphate-glucosyl phosphotransferase initiating biosynthesis of Rhizobium leguminosarum exopolysaccharide. Russ. J. Bioorganic Chem. 2007, 33, 150–155. [Google Scholar] [CrossRef]
- Sheikh, M.O.; Halmo, S.M.; Patel, S.; Middleton, D.; Takeuchi, H.; Schafer, C.M.; West, C.M.; Haltiwanger, R.S.; Avci, F.Y.; Moremen, K.W.; et al. Rapid screening of sugar-nucleotide donor specificities of putative glycosyltransferases. Glycobiology 2017, 27, 206–212. [Google Scholar] [CrossRef] [Green Version]
- Marczak, M.; Dźwierzyńska, M.; Skorupska, A. Homo- and heterotypic interactions between Pss proteins involved in the exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii. Biol. Chem. 2013, 394, 541–559. [Google Scholar] [CrossRef]
- Marczak, M.; Matysiak, P.; Kutkowska, J.; Skorupska, A. PssP2 is a polysaccharide co-polymerase involved in exopolysaccharide chain-length determination in Rhizobium leguminosarum. PLoS ONE 2014, 9, e109106. [Google Scholar] [CrossRef] [Green Version]
- Bathke, J.; Konzer, A.; Remes, B.; McIntosh, M.; Klug, G. Comparative analyses of the variation of the transcriptome and proteome of Rhodobacter sphaeroides throughout growth. BMC Genom. 2019, 20, 358. [Google Scholar] [CrossRef] [Green Version]
- Hosie, A.H.; Allaway, D.; Galloway, C.S.; Dunsby, H.A.; Poole, P.S. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J. Bacteriol. 2002, 184, 4071–4080. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, M.; Nordeste, R.; Doxey, A.C.; Charles, T.C. Transcriptome Analysis of Polyhydroxybutyrate Cycle Mutants Reveals Discrete Loci Connecting Nitrogen Utilization and Carbon Storage in Sinorhizobium meliloti. mSystems 2017, 2, e00035-17. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Han, Q.; Tan, Y.; Ding, H.; Li, J. Current Advances on Structure-Function Relationships of Pyridoxal 5’-Phosphate-Dependent Enzymes. Front. Mol. Biosci. 2019, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Doucette, C.D.; Schwab, D.J.; Wingreen, N.S.; Rabinowitz, J.D. α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 2011, 7, 894–901. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.X.; Rainey, P.B. Genetic analysis of the histidine utilization (hut) genes in Pseudomonas fluorescens SBW25. Genetics 2007, 176, 2165–2176. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Hu, A.; Chen, X.; Luo, S.; Zou, Q.; Xie, J.; He, D.; Li, X.; Cheng, G. Rhizobium leguminosarum Glutathione Peroxidase Is Essential for Oxidative Stress Resistance and Efficient Nodulation. Front. Microbiol. 2021, 12, 627562. [Google Scholar] [CrossRef] [PubMed]
- Aroney, S.T.N.; Poole, P.S.; Sánchez-Cañizares, C. Rhizobial Chemotaxis and Motility Systems at Work in the Soil. Front. Plant Sci. 2021, 12, 725338. [Google Scholar] [CrossRef] [PubMed]
- Kehry, M.R.; Dahlquist, F.W. The methyl-accepting chemotaxis proteins of Escherichia coli. Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J. Biol. Chem. 1982, 257, 10378–10386. [Google Scholar] [CrossRef]
- Compton, K.K.; Scharf, B.E. Rhizobial Chemoattractants, the Taste and Preferences of Legume Symbionts. Front. Plant Sci. 2021, 12, 686465. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Fuentes-Romero, F.; Ruiz-Sainz, J.E.; Janczarek, M.; Vinardell, J.M. Rhizobial Exopolysaccharides: Genetic Regulation of Their Synthesis and Relevance in Symbiosis with Legumes. Int. J. Mol. Sci. 2021, 22, 6233. [Google Scholar] [CrossRef] [PubMed]
- Krol, E.; Klaner, C.; Gnau, P.; Kaever, V.; Essen, L.O.; Becker, A. Cyclic mononucleotide- and Clr-dependent gene regulation in Sinorhizobium meliloti. Microbiology 2016, 162, 1840–1856. [Google Scholar] [CrossRef] [PubMed]
- Nachin, L.; Nannmark, U.; Nyström, T. Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kvint, K.; Nachin, L.; Diez, A.; Nyström, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Siegele, D.A. Universal stress proteins in Escherichia coli. J. Bacteriol. 2005, 187, 6253–6254. [Google Scholar] [CrossRef] [Green Version]
- Chandravanshi, M.; Gogoi, P.; Kanaujia, S.P. Computational characterization of TTHA0379: A potential glycerophosphocholine binding protein of Ugp ATP-binding cassette transporter. Gene 2016, 592, 260–268. [Google Scholar] [CrossRef]
- Rahman, M.M.; Machuca, M.A.; Khan, M.F.; Barlow, C.K.; Schittenhelm, R.B.; Roujeinikova, A. Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori. J. Bacteriol. 2019, 201, e00400-19. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, D.C. Calcium signalling in bacteria. Mol. Microbiol. 2004, 54, 291–297. [Google Scholar] [CrossRef]
- Xi, C.; Schoeters, E.; Vanderleyden, J.; Michiels, J. Symbiosis-specific expression of Rhizobium etli casA encoding a secreted calmodulin-related protein. Proc. Natl. Acad. Sci. USA 2000, 97, 11114–11119. [Google Scholar] [CrossRef] [Green Version]
- Yeats, C.; Bateman, A. The BON domain: A putative membrane-binding domain. Trends Biochem. Sci. 2003, 28, 352–355. [Google Scholar] [CrossRef]
- García Angulo, V.A.; Bonomi, H.R.; Posadas, D.M.; Serer, M.I.; Torres, A.G.; Zorreguieta, A.; Goldbaum, F.A. Identification and characterization of RibN, a novel family of riboflavin transporters from Rhizobium leguminosarum and other proteobacteria. J. Bacteriol. 2013, 195, 4611–4619. [Google Scholar] [CrossRef] [PubMed]
- Trösch, R.; Willmund, F. The conserved theme of ribosome hibernation: From bacteria to chloroplasts of plants. Biol. Chem. 2019, 400, 879–893. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [Green Version]
- Kowalczuk, E.; Lorkiewicz, Z. Transfer of RP4 and R68.45 factors to Rhizobium. Acta Microbiol. Pol. 1979, 28, 221–229. [Google Scholar] [PubMed]
- Dobson, L.; Reményi, I.; Tusnády, G.E. CCTOP: A Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015, 43, W408–W412. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Reeve, W.; Tiwari, R.; Melino, V.; De Meyer, S.; Poole, P.S. Specialised genetic techniques for rhizobia. In Working with rhizobia; Howieson, J., Dilworth, M.J., Eds.; Australian Centre for International Agricultural Research: Canberra, Australia, 2016; pp. 245–282. [Google Scholar]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [Green Version]
- Garg, B.; Dogra, R.C.; Sharma, P.K. High-efficiency transformation of Rhizobium leguminosarum by electroporation. Appl. Environ. Microbiol. 1999, 65, 2802–2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Apicella, M.A. Isolation and characterization of lipopolysaccharides. Methods Mol. Biol. 2008, 431, 3–13. [Google Scholar] [CrossRef]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Manoil, C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991, 34, 61–75. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Sequeira, J.C.; Rocha, M.; Alves, M.M.; Salvador, A.F. UPIMAPI, reCOGnizer and KEGGCharter: Bioinformatics tools for functional annotation and visualization of (meta)-omics datasets. Comput. Struct. Biotechnol. J. 2022, 20, 1798–1810. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2022, 31, 47–53. [Google Scholar] [CrossRef]


| Fusion | Avg PhoA 1 | Avg LacZ 1 | PhoA (%) 2 | LacZ (%) 2 | NAR (PhoA:LacZ) 3 | Location 4 |
|---|---|---|---|---|---|---|
| A85 | 1195.4 | 166.0 | 100 | 20 | 5 | tm |
| A180 | 254.4 | 467.3 | 21 | 56 | 0.375 | tm-c |
| A263 | 140.5 | 835.4 | 12 | 100 | 0.120 | tm-c |
| T201 | 873.5 | 0 | 100 | 0 | >100 | p |
| T243 | 11.6 | 684.2 | 0.01 | 100 | <0.01 | c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marczak, M.; Żebracki, K.; Koper, P.; Horbowicz, A.; Wójcik, M.; Mazur, A. A New Face of the Old Gene: Deletion of the PssA, Encoding Monotopic Inner Membrane Phosphoglycosyl Transferase in Rhizobium leguminosarum, Leads to Diverse Phenotypes That Could Be Attributable to Downstream Effects of the Lack of Exopolysaccharide. Int. J. Mol. Sci. 2023, 24, 1035. https://doi.org/10.3390/ijms24021035
Marczak M, Żebracki K, Koper P, Horbowicz A, Wójcik M, Mazur A. A New Face of the Old Gene: Deletion of the PssA, Encoding Monotopic Inner Membrane Phosphoglycosyl Transferase in Rhizobium leguminosarum, Leads to Diverse Phenotypes That Could Be Attributable to Downstream Effects of the Lack of Exopolysaccharide. International Journal of Molecular Sciences. 2023; 24(2):1035. https://doi.org/10.3390/ijms24021035
Chicago/Turabian StyleMarczak, Małgorzata, Kamil Żebracki, Piotr Koper, Aleksandra Horbowicz, Magdalena Wójcik, and Andrzej Mazur. 2023. "A New Face of the Old Gene: Deletion of the PssA, Encoding Monotopic Inner Membrane Phosphoglycosyl Transferase in Rhizobium leguminosarum, Leads to Diverse Phenotypes That Could Be Attributable to Downstream Effects of the Lack of Exopolysaccharide" International Journal of Molecular Sciences 24, no. 2: 1035. https://doi.org/10.3390/ijms24021035






