Transcriptomic Profiling of Tomato Leaves Identifies Novel Transcription Factors Responding to Dehydration Stress
Abstract
:1. Introduction
2. Results
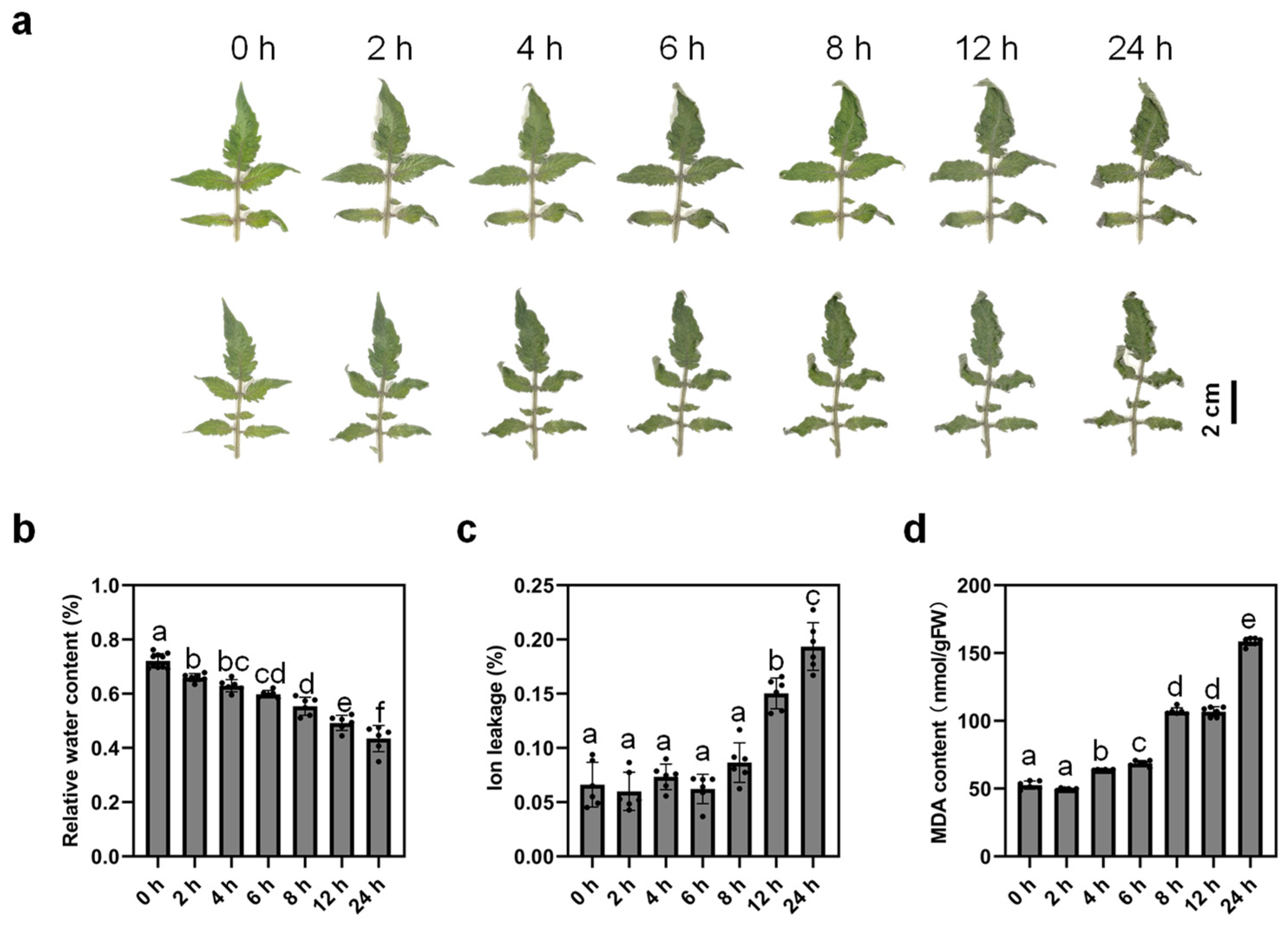
2.1. Dehydration Stress Has a Major Impact on Tomato Leaves
2.2. Dehydration Induces Oxidative Stress Responses
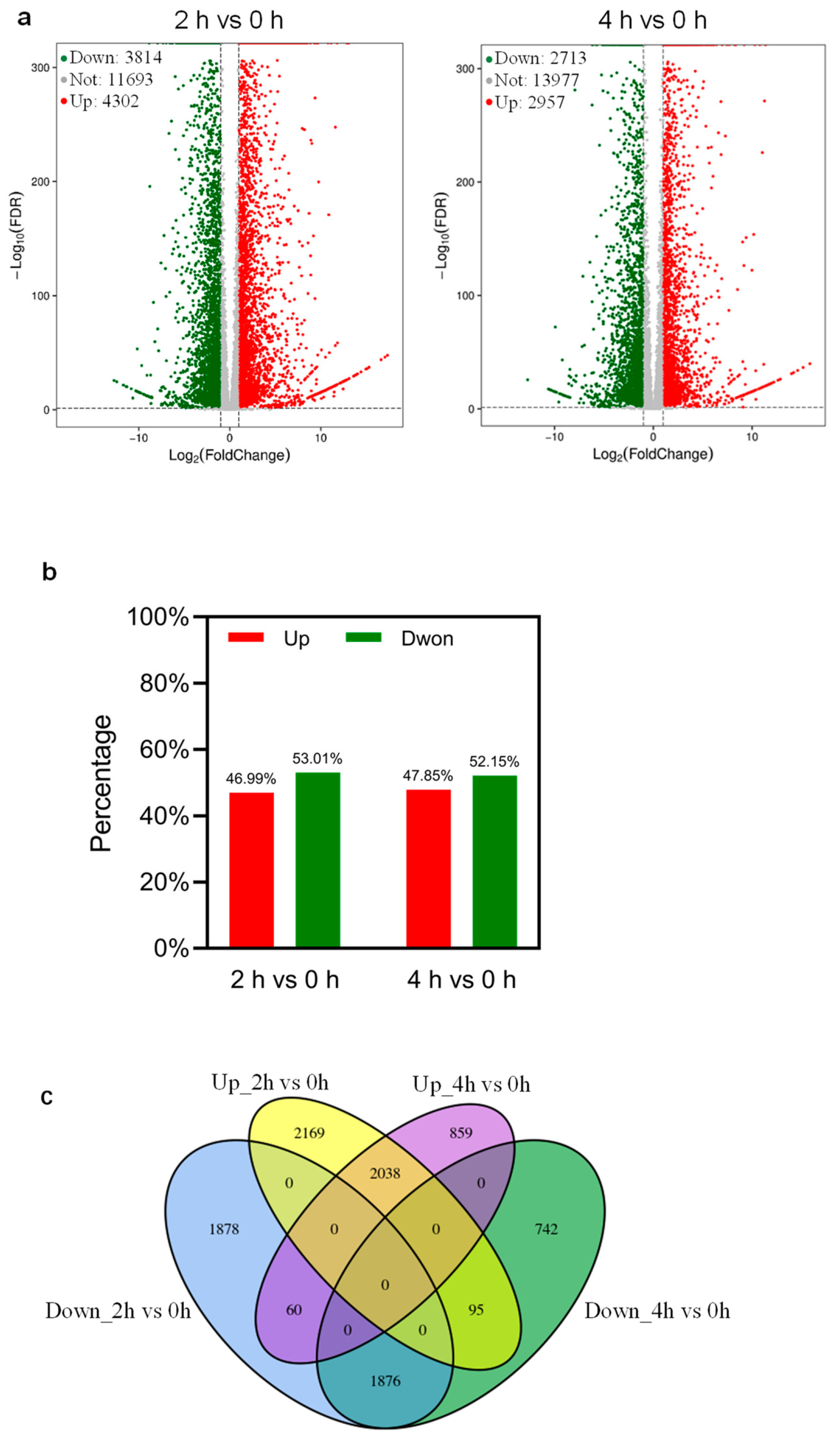
2.3. Transcriptome Analysis Reveals DEGs in Response to Dehydration
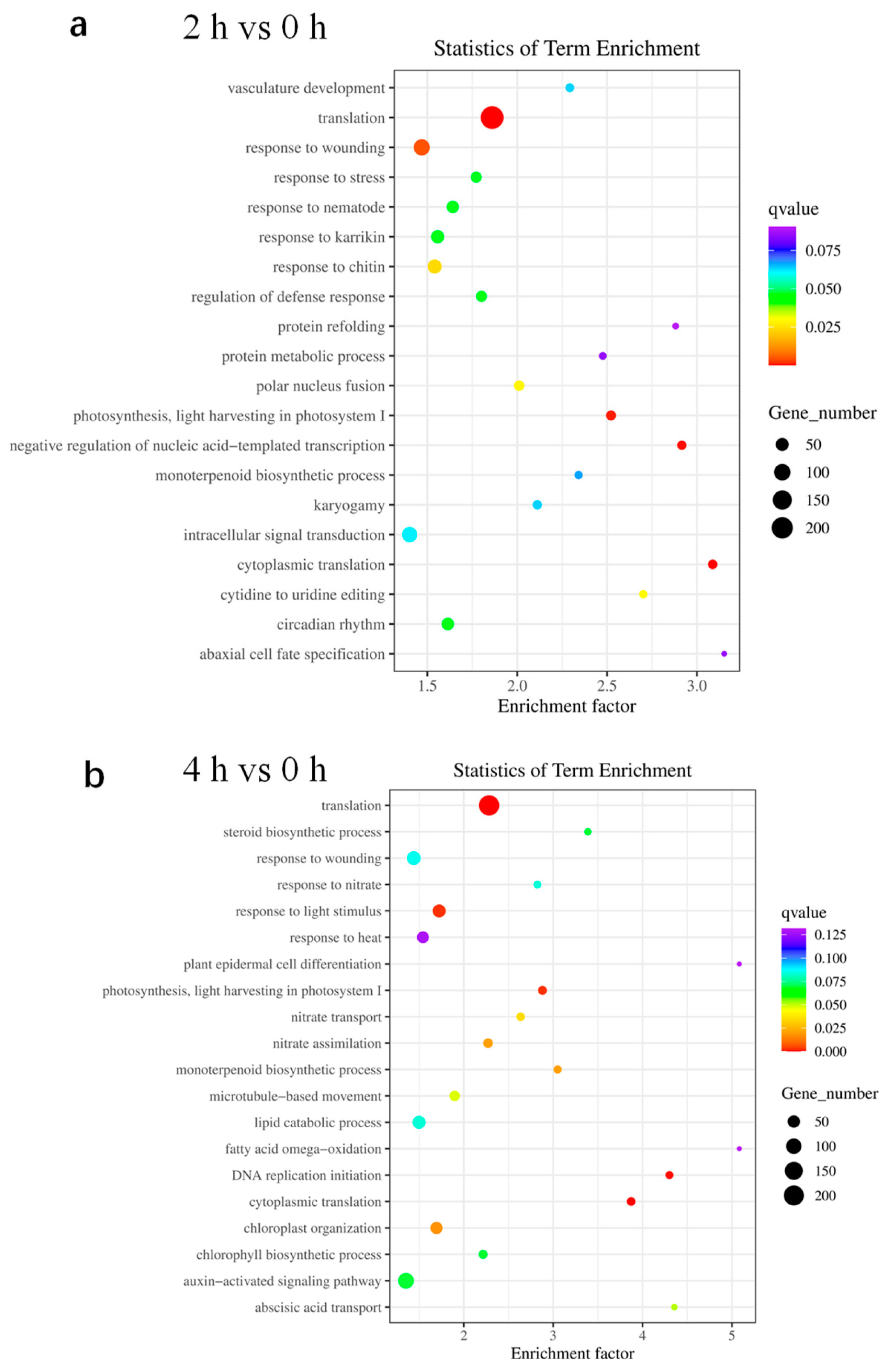
2.4. GO Analysis Reveals Genes Involved in Translation and Stress Response
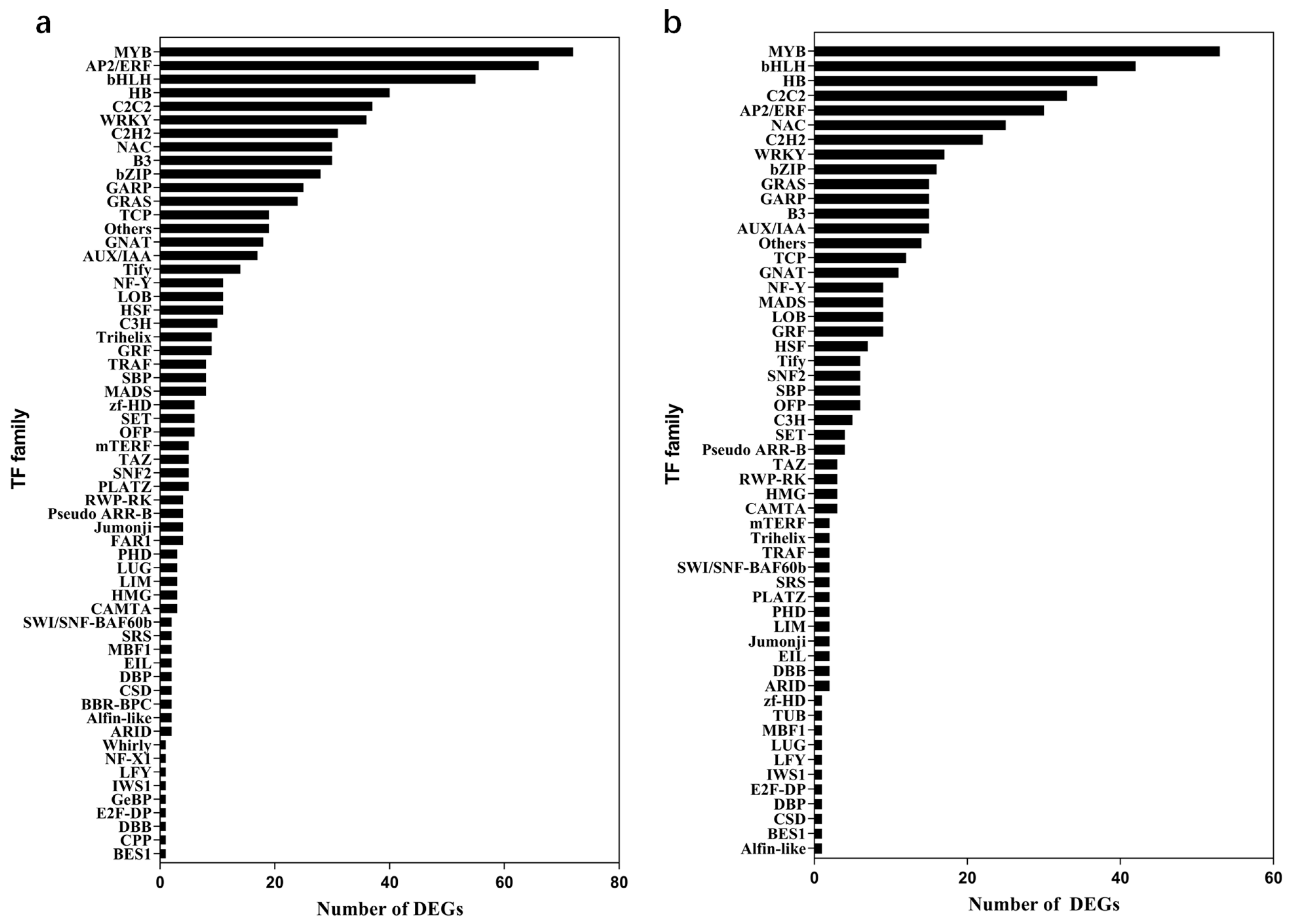
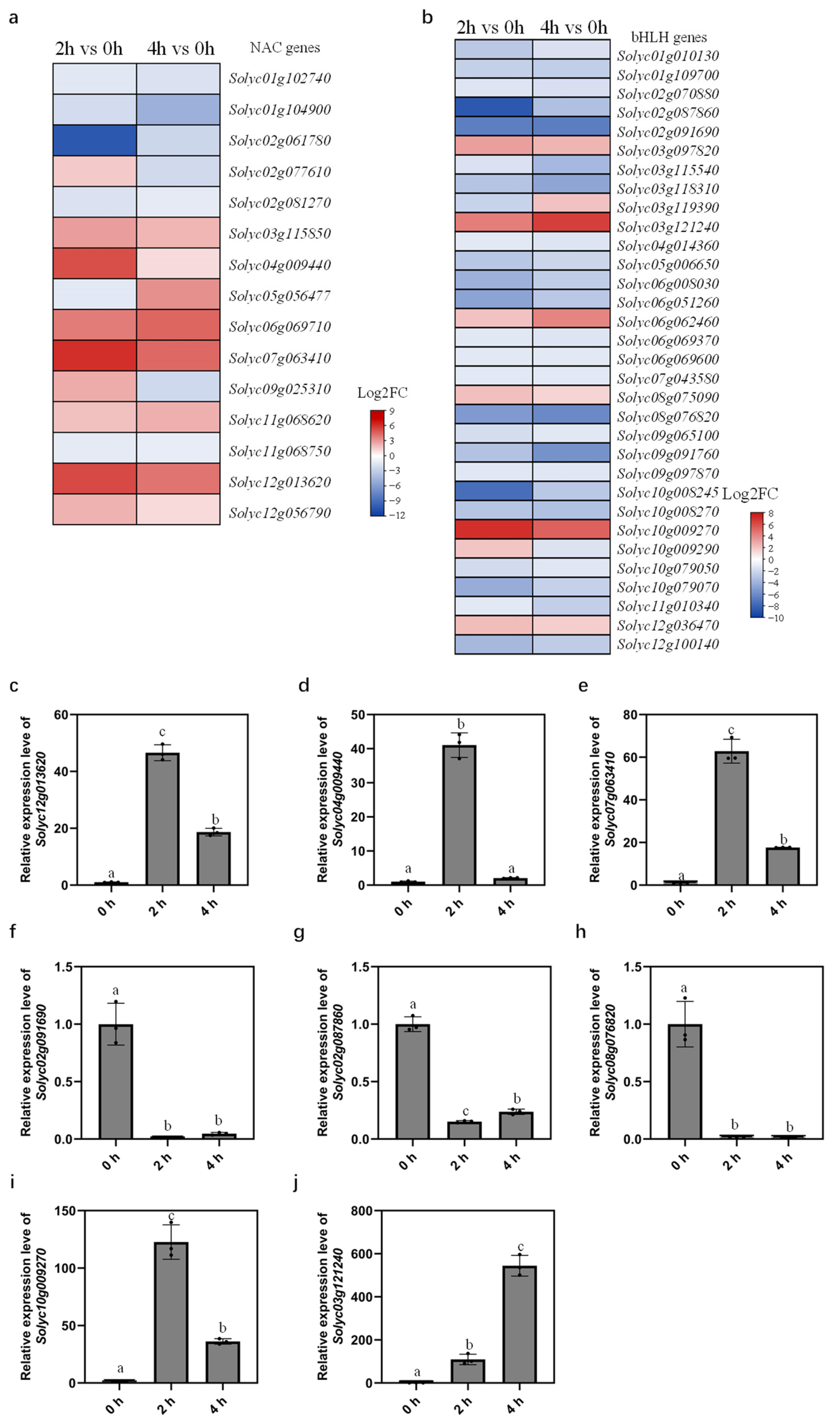
2.5. TF Families Respond to Dehydration Differentially
2.6. qRT-PCR Analysis Validates Expression Patterns of DEGs
3. Discussion
3.1. Comparison of the Effects of Dehydration and Drought Stress on Tomato Leaves
3.2. Dynamic Changes of ROS Levels under Dehydration Conditions
3.3. Transcriptome Profiling Identifies Dehydration-Responsive Genes
3.4. Differentially Expressed TFs
4. Methods and Plant Materials
4.1. Plant Material and Growth Conditions
4.2. Determination of Relative Water Content
4.3. Determination of Ion Leakage
4.4. Enzyme Measurements
4.5. Quantification of H2O2, O2−, and Malondialdehyde (MDA)
4.6. Total RNA Extraction
4.7. Library Construction and RNA-Seq Data Analysis
4.8. Gene Ontology (GO) Enrichment Analysis of Differentially Expressed Genes (DEGs)
4.9. Quantitative Real-Time PCR (qRT-PCR)
4.10. Data Presentation and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Yu, X.; Ottosen, C.O.; Rosenqvist, E.; Zhao, L.; Wang, Y.; Yu, W.; Zhao, T.; Wu, Z. Drought stress had a predominant effect over heat stress on three tomato cultivars subjected to combined stress. BMC Plant Biol. 2017, 17, 24. [Google Scholar] [CrossRef] [Green Version]
- Kamanga, R.M.; Mbega, E.R.; Ndakidemi, P. Drought tolerance mechanisms in plants: Physiological responses associated with water deficit stress in Solanum lycopersicum. Adv. Crop Sci. Technol. 2018, 6, 362. [Google Scholar] [CrossRef]
- Xiong, J.; Zhang, L.; Fu, G.; Yang, Y.; Zhu, C.; Tao, L. Drought-induced proline accumulation is uninvolved with increased nitric oxide, which alleviates drought stress by decreasing transpiration in rice. J. Plant Res. 2012, 125, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Zupin, M.; Sedlar, A.; Kidric, M.; Meglic, V. Drought-induced expression of aquaporin genes in leaves of two common bean cultivars differing in tolerance to drought stress. J. Plant Res. 2017, 130, 735–745. [Google Scholar] [CrossRef]
- Habibi, G.; Ajory, N. The effect of drought on photosynthetic plasticity in Marrubium vulgare plants growing at low and high altitudes. J. Plant Res. 2015, 128, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.T.; Liu, J.H.; Zhang, J.M.; Guo, J. Effects of drought stress on photosynthetic and physiological parameters of tomato. J. Am. Soc. Hortic. Sci. 2020, 145, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Raja, V.; Qadir, S.U.; Alyemeni, M.N.; Ahmad, P. Impact of drought and heat stress individually and in combination on physio-biochemical parameters, antioxidant responses, and gene expression in Solanum lycopersicum. 3 Biotech 2020, 10, 208. [Google Scholar] [CrossRef]
- Ebrahimian-Motlagh, S.; Ribone, P.A.; Thirumalaikumar, V.P.; Allu, A.D.; Chan, R.L.; Mueller-Roeber, B.; Balazadeh, S. JUNGBRUNNEN1 confers drought tolerance downstream of the HD-Zip I transcription factor AtHB13. Front. Plant Sci. 2017, 8, 2118. [Google Scholar] [CrossRef] [Green Version]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef] [Green Version]
- Syed, B.A.; Patel, M.; Patel, A.; Gami, B.; Patel, B. Regulation of antioxidant enzymes and osmo-protectant molecules by salt and drought responsive genes in Bambusa balcooa. J. Plant Res. 2021, 134, 165–175. [Google Scholar] [CrossRef]
- Ilyas, M.; Nisar, M.; Khan, N.; Hazrat, A.; Khan, A.H.; Hayat, K.; Fahad, S.; Khan, A.; Ullah, A. Drought tolerance strategies in plants: A mechanistic approach. J. Plant Growth Regul. 2021, 40, 926–944. [Google Scholar] [CrossRef]
- Yang, L.; Bu, S.; Zhao, S.; Wang, N.; Xiao, J.; He, F.; Gao, X. Transcriptome and physiological analysis of increase in drought stress tolerance by melatonin in tomato. PLoS ONE 2022, 17, e0267594. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.M.; Sonah, H.; et al. Mutation breeding in tomato: Advances, applicability and challenges. Plants 2019, 8, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Du, X. Transcriptome analysis reveals the regulation of cyclic nucleotide-gated ion channels in response to exogenous abscisic acid and calcium treatment under drought stress in tomato. Front. Genet. 2023, 14, 1139087. [Google Scholar] [CrossRef]
- Ye, J.; Wang, X.; Wang, W.; Yu, H.; Ai, G.; Li, C.; Sun, P.; Wang, X.; Li, H.; Ouyang, B.; et al. Genome-wide association study reveals the genetic architecture of 27 agronomic traits in tomato. Plant Physiol. 2021, 186, 2078–2092. [Google Scholar] [CrossRef]
- Wu, Z.; Fan, Y.; Qiu, Y.; Hao, X.; Li, S.; Kang, S. Response of yield and quality of greenhouse tomatoes to water and salt stresses and biochar addition in Northwest China. Agric. Water Manag. 2022, 270, 107736. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef]
- Orellana, S.; Yanez, M.; Espinoza, A.; Verdugo, I.; Gonzalez, E.; Ruiz-Lara, S.; Casaretto, J.A. The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 2010, 33, 2191–2208. [Google Scholar] [CrossRef]
- Nir, I.; Shohat, H.; Panizel, I.; Olszewski, N.; Aharoni, A.; Weiss, D. The tomato DELLA protein PROCERA acts in guard cells to promote stomatal closure. Plant Cell 2017, 29, 3186–3197. [Google Scholar] [CrossRef] [Green Version]
- Loukehaich, R.; Wang, T.; Ouyang, B.; Ziaf, K.; Li, H.; Zhang, J.; Lu, Y.; Ye, Z. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 2012, 63, 5593–5606. [Google Scholar] [CrossRef] [Green Version]
- Asakura, H.; Yamakawa, T.; Tamura, T.; Ueda, R.; Taira, S.; Saito, Y.; Abe, K.; Asakura, T. Transcriptomic and metabolomic analyses provide insights into the upregulation of fatty acid and phospholipid metabolism in tomato fruit under drought stress. J. Agric. Food Chem. 2021, 69, 2894–2905. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Gantait, S.; Azizi, P.; Mazumdar, P. Drought tolerance improvement in Solanum lycopersicum: An insight into “OMICS” approaches and genome editing. 3 Biotech 2022, 12, 63. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.; Gao, J.; Wang, B.; Han, H.; Wang, L.; Peng, R.; Yao, Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, C.; Zhang, Y.; Wang, B.; Ran, Q.; Zhang, J. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 2019, 70, 5471–5486. [Google Scholar] [CrossRef]
- Hu, J.T.; Chen, G.P.; Yin, W.C.; Cui, B.L.; Yu, X.H.; Lu, Y.; Hu, Z.L. Silencing of SlHB2 improves drought, salt stress tolerance, and induces stress-related gene expression in tomato. J. Plant Growth Regul. 2017, 36, 578–589. [Google Scholar] [CrossRef]
- Jian, W.; Zheng, Y.; Yu, T.; Cao, H.; Chen, Y.; Cui, Q.; Xu, C.; Li, Z. SlNAC6, A NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J. Plant Physiol. 2021, 264, 153483. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.; Gao, G. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Meng, X.; Cai, J.; Li, G.; Dong, T.; Li, Z. Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahammed, G.J.; Li, X.; Mao, Q.; Wan, H.; Zhou, G.; Cheng, Y. The SlWRKY81 transcription factor inhibits stomatal closure by attenuating nitric oxide accumulation in the guard cells of tomato under drought. Physiol. Plant. 2021, 172, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Li, X.; Wan, H.J.; Zhou, G.Z.; Cheng, Y. SlWRKY81 reduces drought tolerance by attenuating proline biosynthesis in tomato. Sci. Hortic. 2020, 270, 109444. [Google Scholar] [CrossRef]
- Fuentes-Merlos, M.I.; Bamba, M.; Sato, S.; Higashitani, A. Comparative transcriptome analysis of grafted tomato with drought tolerance. Plants 2022, 11, 1947. [Google Scholar] [CrossRef]
- Qiao, K.; Yao, X.; Zhou, Z.; Xiong, J.; Fang, K.; Lan, J.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase enhanced ABA-mediated drought tolerance in Solanum lycopersicum. J. Plant Physiol. 2023, 280, 153892. [Google Scholar] [CrossRef]
- Huang, K.; Wu, T.; Ma, Z.; Li, Z.; Chen, H.; Zhang, M.; Bian, M.; Bai, H.; Jiang, W.; Du, X. Rice transcription factor OsWRKY55 is involved in the drought response and regulation of plant growth. Int. J. Mol. Sci. 2021, 22, 4337. [Google Scholar] [CrossRef]
- Shim, J.S.; Oh, N.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Kim, J.-K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018, 9, 310. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, L.; Tang, B.; Wu, T.; Chen, G.; Xie, Q.; Hu, Z. Silencing of SlMYB55 affects plant flowering and enhances tolerance to drought and salt stress in tomato. Plant Sci. 2022, 316, 111166. [Google Scholar] [CrossRef]
- Huang, W.; Miao, M.; Kud, J.; Niu, X.; Ouyang, B.; Zhang, J.; Ye, Z.; Kuhl, J.C.; Liu, Y.; Xiao, F. SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytol. 2013, 197, 1214–1224. [Google Scholar] [CrossRef]
- Zhao, P.; Li, Q.; Li, J.; Wang, L.; Ren, Z. Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol. Genet. Genom. 2014, 289, 1183–1207. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Cano-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Tarkowska, D.; Sedaghatmehr, M.; Welsch, M.; Gupta, S.; Mueller-Roeber, B.; Balazadeh, S. The HB40-JUB1 transcriptional regulatory network controls gibberellin homeostasis in Arabidopsis. Mol. Plant 2022, 15, 322–339. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Ida, Y.; Kitajima, S.; Kawazu, T.; Hibino, T.; Hanba, Y.T. Overexpressing the HD-Zip class II transcription factor EcHB1 from Eucalyptus camaldulensis increased the leaf photosynthesis and drought tolerance of Eucalyptus. Sci. Rep. 2019, 9, 14121. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Li, D.H.; Li, H.Y.; Wang, M.C.; Wang, Z.; Liu, J.H. The tomato WRKY transcription factor SLWRKY17 positively regulates drought stress tolerance in transgenic tobacco plants. Russ. J. Plant Physiol. 2023, 69, 154. [Google Scholar] [CrossRef]
- Nikolaeva, M.K.; Maevskaya, S.N.; Shugaev, A.G.; Bukhov, N.G. Effect of drought on chlorophyll content and antioxidant enzyme activities in leaves of three wheat cultivars varying in productivity. Russ. J. Plant Physiol. 2010, 57, 87–95. [Google Scholar] [CrossRef]
- Sakya, A.T.; Sulistyaningsih, E.; Indradewa, D.; Purwanto, B.H. Physiological characters and tomato yield under drought stress. IOP Conf. Ser. Earth Environ. Sci. 2018, 200, 012043. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, X.; Zhao, T.; Ottosen, C.O.; Rosenqvist, E.; Wu, Z. Physiological analysis and transcriptome sequencing reveal the effects of combined cold and drought on tomato leaf. BMC Plant Biol. 2019, 19, 377. [Google Scholar] [CrossRef]
- Babaoğlu Aydaş, S.; Ozturk, S.; Aslım, B. Phenylalanine ammonia lyase (PAL) enzyme activity and antioxidant properties of some cyanobacteria isolates. Food Chem. 2013, 136, 164–169. [Google Scholar] [CrossRef]
- Şirin, S.; Aslım, B. Determination of antioxidant capacity, phenolic acid composition and antiproliferative effect associated with phenylalanine ammonia lyase (PAL) activity in some plants naturally growing under salt stress. Med. Chem. Res. 2019, 28, 229–238. [Google Scholar] [CrossRef]
- Liu, Y.; Wen, L.; Shi, Y.; Su, D.; Lu, W.; Cheng, Y.; Li, Z. Stress-responsive tomato gene SlGRAS4 function in drought stress and abscisic acid signaling. Plant Sci. 2021, 304, 110804. [Google Scholar] [CrossRef]
- Maqsood, H.; Munir, F.; Amir, R.; Gul, A. Genome-wide identification, comprehensive characterization of transcription factors, cis-regulatory elements, protein homology, and protein interaction network of DREB gene family in Solanum lycopersicum. Front. Plant Sci. 2022, 13, 1031679. [Google Scholar] [CrossRef]
- Petrović, I.; Savić, S.; Gricourt, J.; Causse, M.; Jovanović, Z.; Stikić, R. Effect of long-term drought on tomato leaves: The impact on metabolic and antioxidative response. Physiol. Mol. Biol. Plants 2021, 27, 2805–2817. [Google Scholar] [CrossRef]
- Shohat, H.; Eliaz, N.I.; Weiss, D. Gibberellin in tomato: Metabolism, signaling and role in drought responses. Mol. Hortic. 2021, 1, 15. [Google Scholar] [CrossRef]
- Balazadeh, S.; Riaño-Pachón, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10 (Suppl. S1), 63–75. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Balazadeh, S.; Mueller-Roeber, B. Tomato fruit ripening factor NOR controls leaf senescence. J. Exp. Bot. 2019, 70, 2727–2740. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arvidsson, S.; Kwasniewski, M.; Riaño-Pachón, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, S.; Ling, J.; Song, L.; Zhao, L.; Wang, Y.; Zhao, T. Transcriptomic Profiling of Tomato Leaves Identifies Novel Transcription Factors Responding to Dehydration Stress. Int. J. Mol. Sci. 2023, 24, 9725. https://doi.org/10.3390/ijms24119725
Dong S, Ling J, Song L, Zhao L, Wang Y, Zhao T. Transcriptomic Profiling of Tomato Leaves Identifies Novel Transcription Factors Responding to Dehydration Stress. International Journal of Molecular Sciences. 2023; 24(11):9725. https://doi.org/10.3390/ijms24119725
Chicago/Turabian StyleDong, Shuchao, Jiayi Ling, Liuxia Song, Liping Zhao, Yinlei Wang, and Tongmin Zhao. 2023. "Transcriptomic Profiling of Tomato Leaves Identifies Novel Transcription Factors Responding to Dehydration Stress" International Journal of Molecular Sciences 24, no. 11: 9725. https://doi.org/10.3390/ijms24119725







