The Expression of miRNAs Involved in Long-Term Memory Formation in the CNS of the Mollusk Helix lucorum
Abstract
:1. Introduction
2. Results
2.1. Identification of miRNAs Expressed in the CNS of Helix
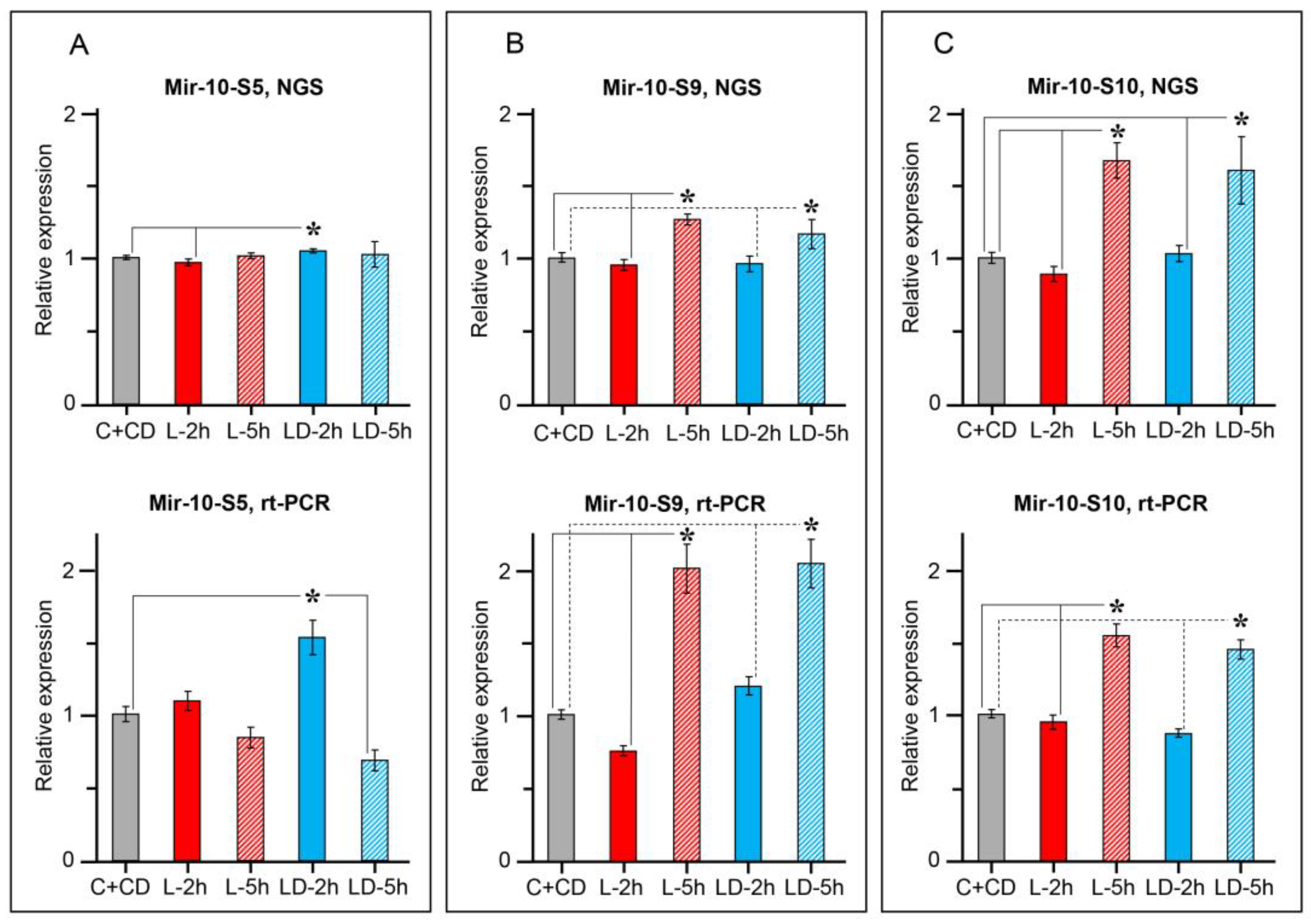
2.2. miRNAs Expression Dynamics in the Helix CNS after Learning
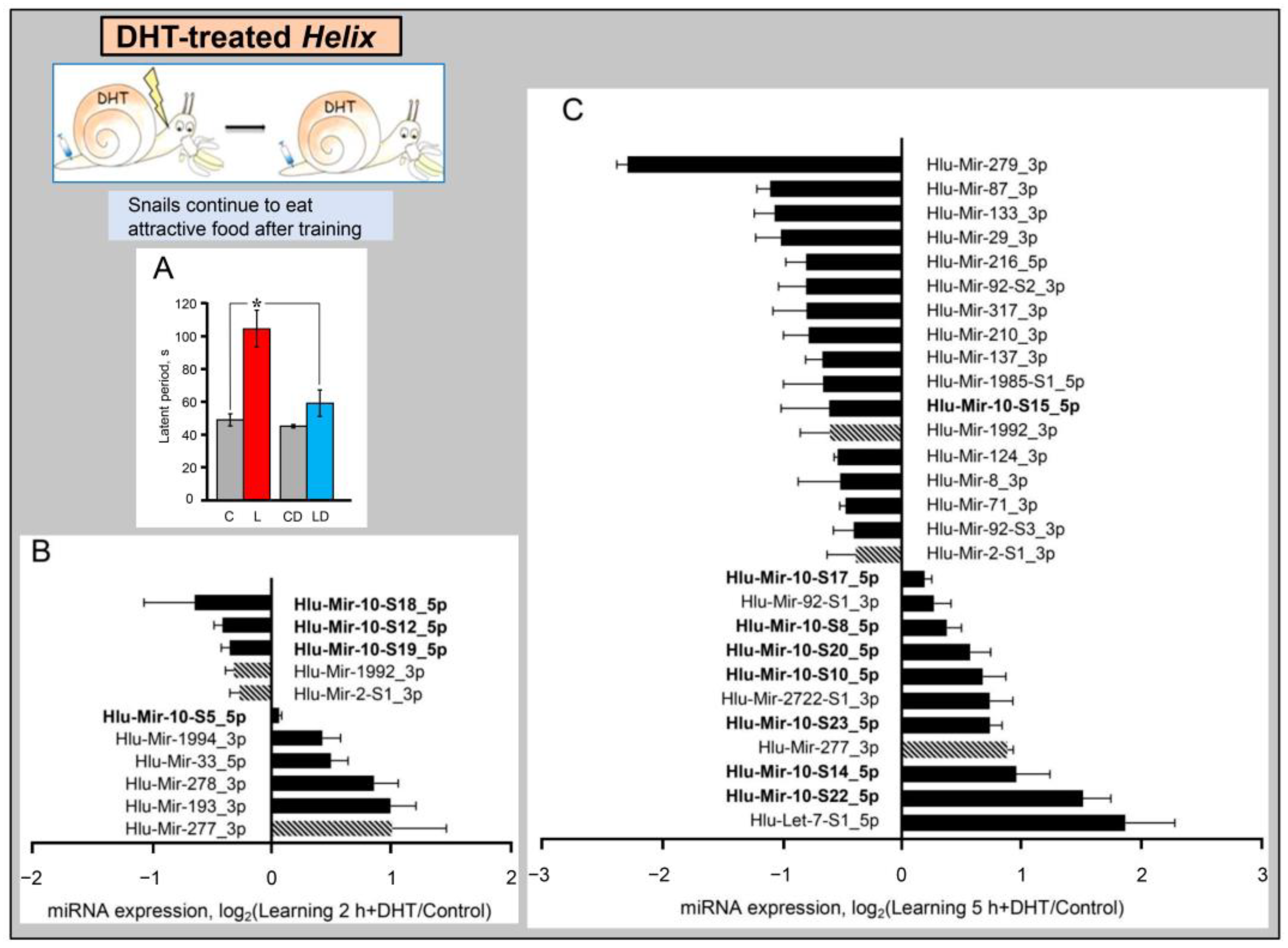
2.3. miRNAs Expression in the CNS of Learning Deficient DHT-Treated Helix after Training
3. Discussion
4. Materials and Methods
4.1. Animals and Conditioned Reflex Formation
4.2. Drugs and the Injection Procedure
4.3. RNA Isolation and Sequencing
4.4. Bioinformatics Analysis
4.5. cDNA Synthesis and Real-Time RT-PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Qin, C. General hallmarks of microRNAs in brain evolution and development. RNA Biol. 2015, 12, 701–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zheng, G.; Dong, D. Coordinated action of histone modification and microRNA regulations in human genome. Gene 2015, 570, 277–281. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Bitetti, A.; Mallory, A.C.; Golini, E.; Carrieri, C.; Carreño Gutiérrez, H.; Perlas, E.; Pérez-Rico, Y.A.; Tocchini-Valentini, G.P.; Enright, A.J.; Norton, W.H.J.; et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 2018, 25, 244–251. [Google Scholar] [CrossRef]
- Rosani, U.; Pallavicini, A.; Venier, P. The miRNA biogenesis in marine bivalves. PeerJ 2016, 4, e1763. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.; Choi, H.; Kim, J. Ebf3-miR218 regulation is involved in the development of dopaminergic neurons. Brain Res. 2014, 1587, 23–32. [Google Scholar] [CrossRef]
- Rajasethupathy, P.; Fiumara, F.; Sheridan, R.; Betel, D.; Puthanveettil, S.V.; Russo, J.J.; Sander, C.; Tuschl, T.; Kandel, E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 2009, 63, 803–817. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, W.Y.; Mao, Y.W.; Gräff, J.; Guan, J.S.; Pan, L.; Mak, G.; Kim, D.; Su, S.C.; Tsai, L.H. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 2010, 466, 1105–1109. [Google Scholar] [CrossRef] [Green Version]
- Konopka, W.; Schütz, G.; Kaczmarek, L. The microRNA contribution to learning and memory. Neuroscientist 2011, 17, 468–474. [Google Scholar] [CrossRef]
- Busto, G.U.; Guven-Ozkan, T.; Fulga, T.A.; Van-Vactor, D.; Davis, R.L. MicroRNAs that promote or inhibit memory formation in Drosophila melanogaster. Genetics 2015, 200, 569–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Li, Z. miRNAs in synapse development and synaptic plasticity. Curr. Opin. Neurobiol. 2017, 45, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Ghosh, S.; Malick, R.C.; Patra, B.C.; Das, B.K. Therapeutic applications of zebrafish (Danio rerio) miRNAs linked with human diseases: A prospective review. Gene 2018, 679, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Eivani, M.; Alijanpour, S.; Arefian, E.; Rezayof, A. Corticolimbic analysis of microRNAs and protein expressions in scopolamine-induced memory loss under stress. Neurobiol. Learn. Mem. 2019, 164, 107065. [Google Scholar] [CrossRef]
- Jawaid, A.; Woldemichael, B.T.; Kremer, E.A.; Laferriere, F.; Gaur, N.; Afroz, T.; Polymenidou, M.; Mansuy, I.M. Memory decline and its reversal in aging and neurodegeneration involve miR-183/96/182 biogenesis. Mol. Neurobiol. 2019, 56, 3451–3462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, H.D.; Jo, W.H.; Hoang, N.H.M.; Kim, M.S. In silico identification of the potential molecular mechanisms involved in protective effects of prolactin on motor and memory deficits induced by 1,2-diacetylbenzene in young and old rats. Neurotoxicology 2022, 93, 45–59. [Google Scholar] [CrossRef]
- Zovoilis, A.; Agbemenyah, H.Y.; Agis-Balboa, R.C.; Stilling, R.M.; Edbauer, D.; Rao, P.; Farinelli, L.; Delalle, I.; Schmitt, A.; Falkai, P.; et al. MicroRNA-34c is a novel target to treat dementias. EMBO J. 2011, 30, 4299–4308. [Google Scholar] [CrossRef]
- Danka Mohammed, C.P.; Park, J.S.; Nam, H.G.; Kim, K. MicroRNAs in brain aging. Mech. Ageing Dev. 2017, 168, 3–9. [Google Scholar] [CrossRef]
- Wingo, T.S.; Yang, J.; Fan, W.; Min Canon, S.; Gerasimov, E.S.; Lori, A.; Logsdon, B.; Yao, B.; Seyfried, N.T.; Lah, J.J.; et al. Brain microRNAs associated with late-life depressive symptoms are also associated with cognitive trajectory and dementia. NPJ Genom. Med. 2020, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Serpente, M.; Fenoglio, C.; D’Anca, M.; Arcaro, M.; Sorrentino, F.; Visconte, C.; Arighi, A.; Fumagalli, G.G.; Porretti, L.; Cattaneo, A.; et al. MiRNA profiling in plasma neural-derived small extracellular vesicles from patients with Alzheimer’s disease. Cells 2020, 9, 1443. [Google Scholar] [CrossRef]
- Wallach, T.; Mossmann, Z.J.; Szczepek, M.; Wetzel, M.; Machado, R.; Raden, M.; Miladi, M.; Kleinau, G.; Krüger, C.; Dembny, P.; et al. MicroRNA-100-5p and microRNA-298-5p released from apoptotic cortical neurons are endogenous Toll-like receptor 7/8 ligands that contribute to neurodegeneration. Mol. Neurodegener. 2021, 16, 80. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.Y.; Cali, C.P.; Lee, E.B. RNA metabolism in neurodegenerative disease. Dis. Model. Mech. 2017, 10, 509–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human microRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandel, E. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain 2012, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moroz, L.L.; Kohn, A.B. Do different neurons age differently? Direct genome-wide analysis of aging in single identified cholinergic neurons. Front. Aging Neurosci. 2010, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korneev, S.A.; Vavoulis, D.V.; Naskar, S.; Dyakonova, V.E.; Kemenes, I.; Kemenes, G. A CREB2-targeting microRNA is required for long-term memory after single-trial learning. Sci. Rep. 2018, 8, 3950. [Google Scholar] [CrossRef] [Green Version]
- Nikitin, V.P.; Solntseva, S.V.; Nikitin, P.V. Protein synthesis inhibitors induce both memory impairment and its recovery. Behav. Brain Res. 2019, 360, 202–208. [Google Scholar] [CrossRef]
- Grinkevich, L.N.; Lisachev, P.D.; Kharchenko, O.A.; Vasil’ev, G.V. Expression of MAP/ERK kinase cascade corresponds to the ability to develop food aversion in terrestrial snail at different stages of ontogenesis. Brain Res. 2008, 1187, 12–19. [Google Scholar] [CrossRef]
- Danilova, A.B.; Kharchenko, O.A.; Shevchenko, K.G.; Grinkevich, L.N. Histone H3 acetylation is asymmetrically induced upon learning in identified neurons of the food aversion network in the mollusk Helix lucorum. Front. Behav. Neurosci. 2010, 4, 1–9. [Google Scholar] [CrossRef]
- Kharchenko, O.A.; Grinkevich, V.V.; Vorobiova, O.V.; Grinkevich, L.N. Learning-induced lateralized activation of the MAPK/ERK cascade in identified neurons of the food-aversion network in the mollusk Helix lucorum. Neurobiol. Learn. Mem. 2010, 94, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Danilova, A.B.; Grinkevich, L.N. Failure of long-term memory formation in juvenile snails is determined by acetylation status of histone H3 and can be improved by NaB treatment. PLoS ONE 2012, 7, e41828. [Google Scholar] [CrossRef] [Green Version]
- Balaban, P. Cellular mechanisms of behavioral plasticity in terrestrial snail. Neurosci. Biobehav. Rev. 2002, 26, 597–630. [Google Scholar] [CrossRef] [PubMed]
- Kandel, E. The molecular biology of memory storage: A dialogue between genes and synapses. Science 2001, 294, 1030–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zakharov, I.S.; Balaban, P.M. Neural mechanisms of age-dependent changes in avoidance behaviour of the snail Helix lucorum. Neuroscience 1987, 23, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Balaban, P.M.; Maksimova, O.A.; Bravarenko, N.I. Behavioral plasticity in snail and its neural mechanisms. Neurosci. Behav. Physiol. 1994, 24, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Grinkevich, L.N.; Lisachev, P.D.; Baranova, K.A.; Kharchenko, O.A. Comparative analysis of MAP/ERK_kinase activation in the CNS of animals with different capability for learning. Ross. Fiziol. Zhurnal Im. IM Sechenova 2006, 92, 536–545. [Google Scholar]
- Grinkevich, L.N.; Vorobiova, O.V. Role of modulatory mediator serotonin in induction of epigenetic processes during long-term memory formation in Helix. Russ. J. Genet. Appl. Res. 2014, 4, 526–532. [Google Scholar] [CrossRef]
- Grinkevich, L.N.; Vorobiova, O.V. Opposing roles of serotonin and neuropeptide FMRFamide in the regulation of epigenetic processes involved in the long-term memory formation. Russ. J. Genet. Appl. Res. 2017, 7, 273–280. [Google Scholar] [CrossRef]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Thatcher, E.J.; Olena, A.F.; Cha, D.J.; Perdigoto, A.L.; Marshall, A.F.; Carter, B.D.; Broadie, K.; Patton, J.G. miR-153 regulates SNAP-25, synaptic transmission, and neuronal development. PLoS ONE 2013, 8, e57080. [Google Scholar] [CrossRef] [PubMed]
- Fiumara, F.; Rajasethupathy, P.; Antonov, I.; Kosmidis, S.; Sossin, W.S.; Kandel, E.R. MicroRNA-22 gates long-term heterosynaptic plasticity in Aplysia through presynaptic regulation of CPEB and downstream targets. Cell Rep. 2015, 11, 1866–1875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, R.S.; Tatarakis, A.; Rudenko, A.; Johnson-Venkatesh, E.M.; Yang, Y.J.; Murphy, E.A.; Todd, T.P.; Schepers, S.T.; Siuti, N.; Martorell, A.J.; et al. A microRNA negative feedback loop downregulates vesicle transport and inhibits fear memory. eLife 2016, 5, e22467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta-Agarwal, S.; Franklin, A.V.; Deramus, T.; Wheelock, M.; Davis, R.L.; McMahon, L.L.; Lubin, F.D. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J. Neurosci. 2012, 32, 5440–5453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jovasevic, V.; Corcoran, K.A.; Leaderbrand, K.; Yamawaki, N.; Guedea, A.L.; Chen, H.J.; Shepherd, G.M.; Radulovic, J. GABAergic mechanisms regulated by miR-33 encode state-dependent fear. Nat. Neurosci. 2015, 18, 1265–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegert, S.; Seo, J.; Kwon, E.J.; Rudenko, A.; Cho, S.; Wang, W.; Flood, Z.; Martorell, A.J.; Ericsson, M.; Mungenast, A.E.; et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015, 18, 1008–1016. [Google Scholar] [CrossRef] [Green Version]
- Malmevik, J.; Petri, R.; Knauff, P.; Brattås, P.L.; Åkerblom, M.; Jakobsson, J. Distinct cognitive effects and underlying transcriptome changes upon inhibition of individual miRNAs in hippocampal neurons. Sci. Rep. 2016, 6, 19879. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Wang, X.; Cai, J.; Du, X.; Sun, H.; Zhang, N. MiR-22-3p regulates amyloid β deposit in mice model of Alzheimer’s disease by targeting mitogen-activated protein kinase 14. Curr. Neurovasc. Res. 2019, 16, 473–480. [Google Scholar] [CrossRef]
- You, Y.H.; Qin, Z.Q.; Zhang, H.L.; Yuan, Z.H.; Yu, X. MicroRNA-153 promotes brain-derived neurotrophic factor and hippocampal neuron proliferation to alleviate autism symptoms through inhibition of JAK-STAT pathway by LEPR. Biosci. Rep. 2019, 39, BSR20181904. [Google Scholar] [CrossRef] [Green Version]
- Balaban, P.M.; Vehovsky, A.; Maksimova, O.A.; Zakharov, I.S. Effect of 5,7-dihydroxytryptamine on the food-aversive conditioning in the snail Helix lucorum L. Brain Res. 1987, 404, 2001–2010. [Google Scholar] [CrossRef]
- Muñoz-Llanos, M.; García-Pérez, M.A.; Xu, X.; Tejos-Bravo, M.; Vidal, E.A.; Moyano, T.C.; Gutiérrez, R.A.; Aguayo, F.I.; Pacheco, A.; García-Rojo, G.; et al. MicroRNA profiling and bioinformatics target analysis in dorsal hippocampus of chronically stressed rats: Relevance to depression pathophysiology. Front. Mol. Neurosci. 2018, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.L.; Zhang, S.; Zhao, H.M.; Xia, S.N.; Jin, Z.; Xu, Y.; Yang, L.; Qu, Y.; Huang, S.Y.; Duan, M.J.; et al. MicroRNA-153 impairs presynaptic plasticity by blocking vesicle release following chronic brain hypoperfusion. Cell Commun. Signal 2020, 18, 57. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, A.; Constantine-Paton, M. Postsynaptic BDNF-TrkB signaling in synapse maturation, plasticity, and disease. Dev. Neurobiol. 2010, 70, 304–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muddashetty, R.S.; Nalavadi, V.C.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.T.; Bassell, G.J. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell 2011, 42, 673–688. [Google Scholar] [CrossRef] [Green Version]
- Lukiw, W.J.; Alexandrov, P.N. Regulation of complement factor H (CFH) by multiple miRNAs in Alzheimer’s disease (AD) brain. Mol. Neurobiol. 2012, 46, 11–19. [Google Scholar] [CrossRef]
- Varendi, K.; Kumar, A.; Härma, M.A.; Andressoo, J.O. miR-1, miR-10b, miR-155, and miR-191 are novel regulators of BDNF. Cell. Mol. Life Sci. 2014, 71, 4443–4456. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Zhu, J. Effects of sleep deprivation on behaviors and abnormal hippocampal BDNF/miR-10B expression in rats with chronic stress depression. Int. J. Clin. Exp. Pathol. 2015, 8, 586–593. [Google Scholar]
- Wang, Y.; Wang, B.; Shao, X.; Liu, M.; Jiang, K.; Wang, M.; Wang, L. Identification and profiling of microRNAs during embryogenesis in the red claw crayfish Cherax quadricarinatus. Front. Physiol. 2020, 11, 878. [Google Scholar] [CrossRef]
- Fromm, B.; Domanska, D.; Høye, E.; Ovchinnikov, V.; Kang, W.; Aparicio-Puerta, E.; Johansen, M.; Flatmark, K.; Mathelier, A.; Hovig, E.; et al. MirGeneDB 2.0: The metazoan microRNA complement. Nucleic Acids Res. 2020, 48, D132–D141. [Google Scholar] [CrossRef] [Green Version]
- Ameres, S.L.; Horwich, M.D.; Hung, J.H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010, 328, 1534–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Wolter, J.M.; Le, H.H.; Linse, A.; Godlove, V.A.; Nguyen, T.D.; Kotagama, K.; Lynch, A.; Rawls, A.; Mangone, M. Evolutionary patterns of metazoan microRNAs reveal targeting principles in the let-7 and miR-10 families. Genome Res. 2017, 27, 53–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://mirgenedb.org/browse?org=cgi&seed=GGACGGA (accessed on 20 October 2022).
- Shenoy, A.; Danial, M.; Blelloch, R.H. Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J. 2015, 34, 1180–1194. [Google Scholar] [CrossRef] [Green Version]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.C.; Chawla, G.; Sokol, N. let-7-Complex microRNAs regulate Broad-Z3, which together with Chinmo maintains adult lineage neurons in an immature state. G3 (Bethesda) 2020, 10, 1393–1401. [Google Scholar] [CrossRef] [Green Version]
- Zhai, L.; Li, Y.; Lan, X.; Ai, L. MicroRNA-10a-5p suppresses cancer proliferation and division in human cervical cancer by targeting BDNF. Exp. Ther. Med. 2017, 14, 6147–6151. [Google Scholar] [CrossRef] [Green Version]
- Müller, S. In silico analysis of regulatory networks underlines the role of miR-10b-5p and its target BDNF in huntington’s disease. Transl. Neurodegener. 2014, 3, 17. [Google Scholar] [CrossRef] [Green Version]
- Bai, G.; Ambalavanar, R.; Wei, D.; Dessem, D. Down regulation of selective microRNAs in trigeminal ganglion neurons following inflammatory muscle pain. Mol. Pain 2007, 3, 15. [Google Scholar] [CrossRef] [Green Version]
- Banzhaf-Strathmann, J.; Benito, E.; May, S.; Arzberger, T.; Tahirovic, S.; Kretzschmar, H.; Fischer, A.; Edbauer, D. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 2014, 33, 1667–1680. [Google Scholar] [CrossRef] [Green Version]
- Macedo, L.M.; Nunes, F.M.; Freitas, F.C.; Pires, C.V.; Tanaka, E.D.; Martins, J.R.; Piulachs, M.D.; Cristino, A.S.; Pinheiro, D.G.; Simões, Z.L. MicroRNA signatures characterizing caste-independent ovarian activity in queen and worker honeybees (Apis mellifera L.). Insect Mol. Biol. 2016, 25, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Vetere, G.; Barbato, C.; Pezzola, S.; Frisone, P.; Aceti, M.; Ciotti, M.; Cogoni, C.; Ammassari-Teule, M.; Ruberti, F. Selective inhibition of miR-92 in hippocampal neurons alters contextual fear memory. Hippocampus 2014, 24, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.P.; Singewald, N. Role of MicroRNAs in anxiety and anxiety-related disorders. Curr. Top. Behav. Neurosci. 2019, 42, 185–219. [Google Scholar] [PubMed]
- Letellier, M.; Elramah, S.; Mondin, M.; Soula, A.; Penn, A.; Choquet, D.; Landry, M.; Thoumine, O.; Favereaux, A. miR-92a regulates expression of synaptic GluA1-containing AMPA receptors during homeostatic scaling. Nat. Neurosci. 2014, 17, 1040–1042. [Google Scholar] [CrossRef]
- Behura, S.K.; Whitfield, C.W. Correlated expression patterns of microRNA genes with age-dependent behavioural changes in honeybee. Insect Mol. Biol. 2010, 19, 431–439. [Google Scholar] [CrossRef]
- Kremer, E.A.; Gaur, N.; Lee, M.A.; Engmann, O.; Bohacek, J.; Mansuy, I.M. Interplay between TETs and microRNAs in the adult brain for memory formation. Sci. Rep. 2018, 8, 1678. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhao, L.; Kan, B.; Shi, H.; Han, J. miR-22 exerts anti-alzheimic effects via the regulation of apoptosis of hippocampal neurons. Cell. Mol. Biol. (Noisy-Le-Grand) 2018, 64, 84–89. [Google Scholar] [CrossRef]
- Michely, J.; Kraft, S.; Müller, U. miR-12 and miR-124 contribute to defined early phases of long-lasting and transient memory. Sci. Rep. 2017, 7, 7910. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Shu, X.; Liu, D.; Shang, Y.; Wu, Y.; Pei, L.; Xu, X.; Tian, Q.; Zhang, J.; Qian, K.; et al. EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron 2012, 73, 774–788. [Google Scholar] [CrossRef] [Green Version]
- Grace, P.M.; Strand, K.A.; Galer, E.L.; Maier, S.F.; Watkins, L.R. MicroRNA-124 and microRNA-146a both attenuate persistent neuropathic pain induced by morphine in male rats. Brain Res. 2018, 1692, 9–11. [Google Scholar] [CrossRef]
- Wright, C.; Turner, J.A.; Calhoun, V.D.; Perrone-Bizzozero, N. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet. 2013, 4, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwivedi, Y. MicroRNAs in depression and suicide: Recent insights and future perspectives. J. Affect. Disord. 2018, 240, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Thomas, K.T.; Gross, C.; Bassell, G.J. MicroRNAs sculpt neuronal communication in a tight balance that is lost in neurological disease. Front. Mol. Neurosci. 2018, 11, 455. [Google Scholar] [CrossRef] [PubMed]
- Olde Loohuis, N.F.; Ba, W.; Stoerchel, P.H.; Kos, A.; Jager, A.; Schratt, G.; Martens, G.J.; van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA-137 controls AMPA-receptor-mediated transmission and mGluR-dependent LTD. Cell Rep. 2015, 11, 1876–1884. [Google Scholar] [CrossRef] [Green Version]
- Lippi, G.; Steinert, J.R.; Marczylo, E.L.; D’Oro, S.; Fiore, R.; Forsythe, I.D.; Schratt, G.; Zoli, M.; Nicotera, P.; Young, K.W. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J. Cell Biol. 2011, 194, 889–904. [Google Scholar] [CrossRef]
- Volpicelli, F.; Speranza, L.; Pulcrano, S.; De Gregorio, R.; Crispino, M.; De Sanctis, C.; Leopoldo, M.; Lacivita, E.; di Porzio, U.; Bellenchi, G.C.; et al. The microRNA-29a modulates serotonin 5-HT7 receptor expression and its effects on hippocampal neuronal morphology. Mol. Neurobiol. 2019, 56, 8617–8627. [Google Scholar] [CrossRef]
- Fernandez, F.; Soon, I.; Li, Z.; Kuan, T.C.; Min, D.H.; Wong, E.S.; Demidov, O.N.; Paterson, M.C.; Dawe, G.; Bulavin, D.V.; et al. Wip1 phosphatase positively modulates dendritic spine morphology and memory processes through the p38MAPK signaling pathway. Cell Adh. Migr. 2012, 6, 333–343. [Google Scholar] [CrossRef] [Green Version]
- Grinkevich, L.N. p38 MAPK is involved in regulation of epigenetic mechanisms of food aversion learning. Bull. Exp. Biol. Med. 2017, 163, 412–414. [Google Scholar] [CrossRef]
- Horsham, J.L.; Ganda, C.; Kalinowski, F.C.; Brown, R.A.; Epis, M.R.; Leedman, P.J. MicroRNA-7: A miRNA with expanding roles in development and disease. Int. J. Biochem. Cell Biol. 2015, 69, 215–224. [Google Scholar] [CrossRef]
- Lin, Q.; Ponnusamy, R.; Widagdo, J.; Choi, J.A.; Ge, W.; Probst, C.; Buckley, T.; Lou, M.; Bredy, T.W.; Fanselow, M.S.; et al. MicroRNA-mediated disruption of dendritogenesis during a critical period of development influences cognitive capacity later in life. Proc. Natl. Acad. Sci. USA 2017, 114, 9188–9193. [Google Scholar] [CrossRef] [Green Version]
- Yuva-Aydemir, Y.; Simkin, A.; Gascon, E.; Gao, F.B. MicroRNA-9: Functional evolution of a conserved small regulatory RNA. RNA Biol. 2011, 8, 557–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garaffo, G.; Conte, D.; Provero, P.; Tomaiuolo, D.; Luo, Z.; Pinciroli, P.; Peano, C.; D’Atri, I.; Gitton, Y.; Etzion, T.; et al. The Dlx5 and Foxg1 transcription factors, linked via miRNA-9 and -200, are required for the development of the olfactory and GnRH system. Mol. Cell. Neurosci. 2015, 68, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Niu, J.J.; Zhou, J.F.; Wei, Y.S. MicroRNA-96 is responsible for sevoflurane-induced cognitive dysfunction in neonatal rats via inhibiting IGF1R. Brain Res. Bull. 2019, 144, 140–148. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zhao, X.; Lei, Y.; Nie, J.; Lu, X.; Song, J.; Wang, L.; Li, H.; Liu, F.; Zhang, Y.; et al. Whole-transcriptome analysis of aluminum-exposed rat hippocampus and identification of ceRNA networks to investigate neurotoxicity of Al. Mol. Ther. Nucleic Acids 2021, 26, 1401–1417. [Google Scholar] [CrossRef] [PubMed]
- Woldemichael, B.T.; Jawaid, A.; Kremer, E.A.; Gaur, N.; Krol, J.; Marchais, A.; Mansuy, I.M. The microRNA cluster miR-183/96/182 contributes to long-term memory in a protein phosphatase 1-dependent manner. Nat. Commun. 2016, 7, 12594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreyer, J.L. New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med. 2010, 2, 92. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Wei, Y.; Jiang, F.; Wang, Y.; Guo, X.; He, J.; Kang, L. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLoS Genet. 2014, 10, e1004206. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Li, F.; Zhang, W.; Jia, P. Growth of glioblastoma is inhibited by miR-133-mediated EGFR suppression. Tumour Biol. 2015, 36, 9553–9558. [Google Scholar] [CrossRef]
- Kong, Y.; Wu, J.; Yuan, L. MicroRNA expression analysis of adult-onset Drosophila Alzheimer’s disease model. Curr. Alzheimer Res. 2014, 11, 882–891. [Google Scholar] [CrossRef]
- Marco, A.; Hooks, K.B.; Griffiths-Jones, S. Evolution and function of the extended miR-2 microRNA family. RNA Biol. 2012, 9, 242–248. [Google Scholar] [CrossRef] [Green Version]
- Gaeta, A.L.; Nourse, J.B., Jr.; Willicott, K.; McKay, L.E.; Keogh, C.M.; Peter, K.; Russell, S.N.; Hamamichi, S.; Berkowitz, L.A.; Caldwell, K.A.; et al. Systemic RNA Interference Defective (SID) genes modulate dopaminergic neurodegeneration in C. elegans. PLoS Genet. 2022, 18, e1010115. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Ren, X.; Zheng, Y.; Qian, J.; Xu, L.; Sun, M. MiR-315 is required for neural development and represses the expression of dFMR1 in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2020, 525, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Poidevin, M.; Li, H.; Chen, D.; Jin, P. MicroRNA-277 modulates the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2012, 8, e1002681. [Google Scholar] [CrossRef] [PubMed]
- Esslinger, S.M.; Schwalb, B.; Helfer, S.; Michalik, K.M.; Witte, H.; Maier, K.C.; Martin, D.; Michalke, B.; Tresch, A.; Cramer, P.; et al. Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol. 2013, 10, 1042–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finger, F.; Ottens, F.; Springhorn, A.; Drexel, T.; Proksch, L.; Metz, S.; Cochella, L.; Hoppe, T. Olfaction regulates organismal proteostasis and longevity via microRNA-dependent signaling. Nat. Metab. 2019, 1, 350–359. [Google Scholar] [CrossRef]
- Hsieh, Y.W.; Chang, C.; Chuang, C.F. The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet. 2012, 8, e1002864. [Google Scholar] [CrossRef] [Green Version]
- Kitatani, Y.; Tezuka, A.; Hasegawa, E.; Yanagi, S.; Togashi, K.; Tsuji, M.; Kondo, S.; Parrish, J.Z.; Emoto, K. Drosophila miR-87 promotes dendrite regeneration by targeting the transcriptional repressor Tramtrack69. PLoS Genet. 2020, 16, e1008942. [Google Scholar] [CrossRef]
- Watts, M.E.; Williams, S.M.; Nithianantharajah, J.; Claudianos, C. Hypoxia-induced microRNA-210 targets neurodegenerative pathways. Noncoding RNA 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Song, Y.; Wang, H.; Liu, K.; Shao, Z.; Shang, Z. MiR-210-3p-EphrinA3-PI3K/AKT axis regulates the progression of oral cancer. J. Cell. Mol. Med. 2020, 24, 4011–4022. [Google Scholar] [CrossRef] [Green Version]
- Murai, K.K.; Nguyen., L.N.; Irie, F.; Yu, Y.; Pasquale, E.B. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat. Neurosci. 2003, 6, 153–160. [Google Scholar] [CrossRef]
- Qin, Q.H.; Wang, Z.L.; Tian, L.Q.; Gan, H.Y.; Zhang, S.W.; Zeng, Z.J. The integrative analysis of microRNA and mRNA expression in Apis mellifera following maze-based visual pattern learning. Insect Sci. 2014, 21, 619–636. [Google Scholar] [CrossRef] [PubMed]
- McNeill, E.; Van Vactor, D. MicroRNAs shape the neuronal landscape. Neuron 2012, 75, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Anokhin, K.V. The molecular scenarios of the consolidation of long-term memory. Zhurnal Vyss. Nervn. Deiatelnosti Im. IP Pavlov. 1997, 47, 261–279. [Google Scholar] [PubMed]
- Gantier, M.P.; McCoy, C.E.; Rusinova, I.; Saulep, D.; Wang, D.; Xu, D.; Irving, A.T.; Behlke, M.A.; Hertzog, P.J.; Mackay, F.; et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011, 39, 5692–5703. [Google Scholar] [CrossRef] [Green Version]
- Kye, M.J.; Neveu, P.; Lee, Y.S.; Zhou, M.; Steen, J.A.; Sahin, M.; Kosik, K.S.; Silva, A.J. NMDA mediated contextual conditioning changes miRNA expression. PLoS ONE 2011, 6, e24682. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]

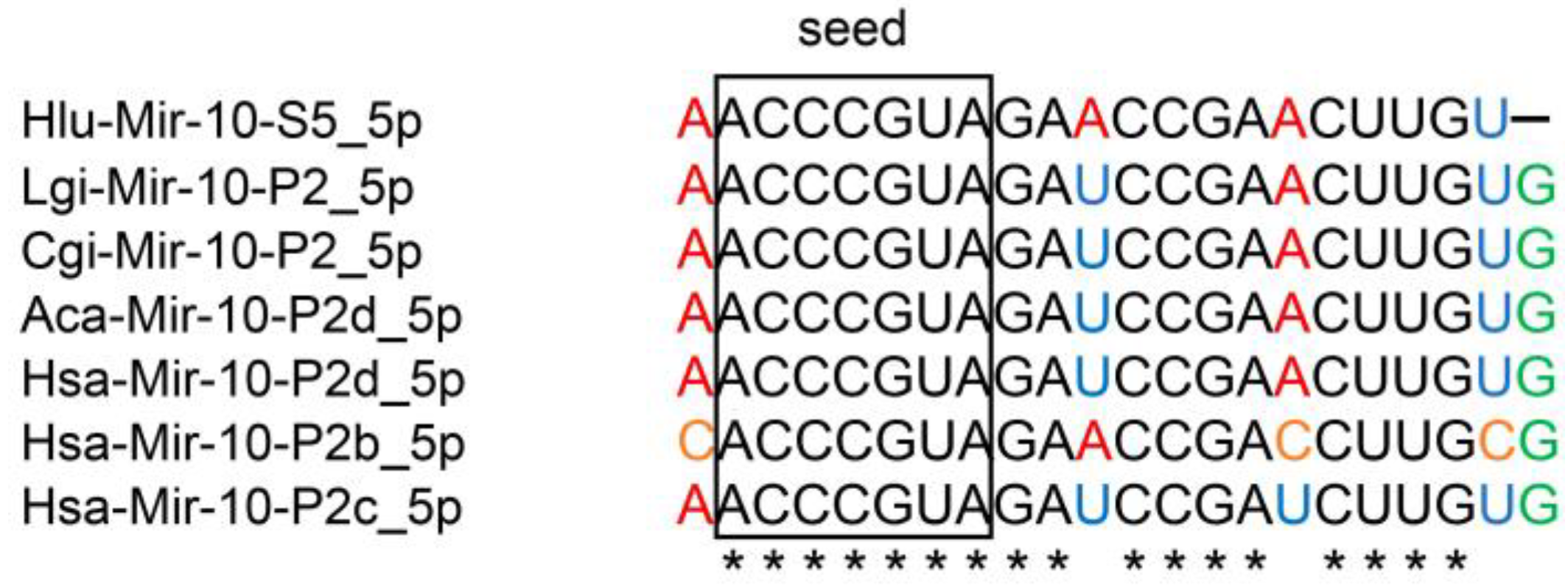


| Subfamily | Seed Region | Helix miRNA | Human Homologue |
|---|---|---|---|
| MIR-10-P1 (mir-10) | ACCCUGU | from hlu-mir-10-S1 to hlu-mir-10-S3 | hsa-mir-10a [Gene ID: 406902] |
| MIR-10-P2 (mir-99/100) | ACCCGUA | from hlu-mir-10-S4 to hlu-mir-10-S7 | hsa-mir-100 [Gene ID: 406892] |
| MIR-10-P3 (mir-125) | CCCUGAG | from hlu-mir-10-S8 to hlu-mir-10-S26 | hsa-mir-125b [Gene ID: 406911] |
| MicroRNA dynamics 2 h after training | |||||
| MicroRNA | Normal animals | DHT treated | MicroRNA | Normal animals | DHT treated |
| Hlu-Mir-2-S1_3p | ⇩ | Hlu-Mir-76-S3_3p | ↑ | ||
| Hlu-Mir-10-S5_5p | ↑ | Hlu-Mir-133_3p | ↑ | ||
| Hlu-Mir-10-S6_5p | ⇩ | Hlu-Mir-193_3p | ↑ | ||
| Hlu-Mir-10-S12_5p | ⇩ | Hlu-Mir-277_3p | ↑ | ||
| Hlu-Mir-10-S18_5p | ⇩ | Hlu-Mir-278_3p | ↑ | ||
| Hlu-Mir-10-S19_5p | ⇩ | Hlu-Mir-1985-S2_3p | ↑ | ||
| Hlu-Mir-33_5p | ↑ | Hlu-Mir-2722-S1_3p | ⇩ | ||
| MicroRNA dynamics 5 h after training | |||||
| MicroRNA | Normal animals | DHT treated | MicroRNA | Normal animals | DHT treated |
| Hlu-Let-7-S1_5p | ↑ | Hlu-Mir-71_3p | ⇩ | ⇩ | |
| Hlu-Mir-2-S1_3p | ⇩ | Hlu-Mir-87_3p | ⇩ | ⇩ | |
| Hlu-Mir-7_5p | ⇩ | Hlu-Mir-92-S1_3p | ↑ | ||
| Hlu-Mir-8_3p | ⇩ | ⇩ | Hlu-Mir-92-S2_3p | ⇩ | ⇩ |
| Hlu-Mir-9_5p | ⇩ | Hlu-Mir-92-S3_3p | ⇩ | ⇩ | |
| Hlu-Mir-10-S8_5p | ↑ | ↑ | Hlu-Mir-96_5p | ↑ | |
| Hlu-Mir-10-S9_5p | ↑ | ↑ | Hlu-Mir-124_3p | ⇩ | ⇩ |
| Hlu-Mir-10-S10_5p | ↑ | ↑ | Hlu-Mir-133_3p | ⇩ | ⇩ |
| Hlu-Mir-10-S14_5p | ↑ | ↑ | Hlu-Mir-137_3p | ⇩ | ⇩ |
| Hlu-Mir-10-S15_5p | ⇩ | ⇩ | Hlu-Mir-153-S1_3p | ⇩ | |
| Hlu-Mir-10-S17_5p | ↑ | ↑ | Hlu-Mir-153-S2_3p | ⇩ | |
| Hlu-Mir-10-S20_5p | ↑ | ↑ | Hlu-Mir-210_3p | ⇩ | ⇩ |
| Hlu-Mir-10-S21_5p | ↑ | Hlu-Mir-216_5p | ⇩ | ⇩ | |
| Hlu-Mir-10-S22_5p | ↑ | ↑ | Hlu-Mir-277_3p | ↑ | |
| Hlu-Mir-10-S23_5p | ↑ | ↑ | Hlu-Mir-279_3p | ⇩ | |
| Hlu-Mir-10-S24_5p | ↑ | Hlu-Mir-317_3p | ⇩ | ||
| Hlu-Mir-10-S25_5p | ↑ | Hlu-Mir-1985-S1_5p | ⇩ | ⇩ | |
| Hlu-Mir-22-S1_3p | ⇩ | Hlu-Mir-1992_3p | ⇩ | ||
| Hlu-Mir-29_3p | ⇩ | Hlu-Mir-2722-S1_3p | ↑ | ↑ | |
| Hlu-Mir-33_5p | ⇩ | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasiliev, G.V.; Ovchinnikov, V.Y.; Lisachev, P.D.; Bondar, N.P.; Grinkevich, L.N. The Expression of miRNAs Involved in Long-Term Memory Formation in the CNS of the Mollusk Helix lucorum. Int. J. Mol. Sci. 2023, 24, 301. https://doi.org/10.3390/ijms24010301
Vasiliev GV, Ovchinnikov VY, Lisachev PD, Bondar NP, Grinkevich LN. The Expression of miRNAs Involved in Long-Term Memory Formation in the CNS of the Mollusk Helix lucorum. International Journal of Molecular Sciences. 2023; 24(1):301. https://doi.org/10.3390/ijms24010301
Chicago/Turabian StyleVasiliev, Gennady V., Vladimir Y. Ovchinnikov, Pavel D. Lisachev, Natalia P. Bondar, and Larisa N. Grinkevich. 2023. "The Expression of miRNAs Involved in Long-Term Memory Formation in the CNS of the Mollusk Helix lucorum" International Journal of Molecular Sciences 24, no. 1: 301. https://doi.org/10.3390/ijms24010301





