Non-Coding RNAs Regulate Spontaneous Abortion: A Global Network and System Perspective
Abstract
:1. Introduction
2. Pathogenesis of SA
3. miRNAs and Spontaneous Abortion
3.1. Overview of miRNAs
3.2. miRNAs Affect SA by Regulating Apoptosis
3.3. miRNAs Are Key Regulators in the Physiological Processes of the Placenta
3.4. miRNAs Participate in the Regulation of Immunity
4. lncRNAs and Spontaneous Abortion
4.1. Overview of lncRNAs
4.2. lncRNAs Regulate Target mRNAs Directly
4.3. lncRNAs Regulate Target miRNAs to Regulate Their Downstream Target mRNAs
5. circRNAs and Spontaneous Abortion
5.1. Overview of circRNAs
5.2. circRNAs and SA
6. ceRNA Network in SA: A Systematic Perspective to Elucidate the Regulation of SA by ncRNAs
6.1. LncRNAs-miRNAs-mRNAs Network
6.2. CircRNAs-miRNAs-mRNAs Network
6.3. lncRNAs/circRNAs-miRNAs-mRNAs Network
7. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Regan, L.; Rai, R. Epidemiology and the medical causes of miscarriage. Baillieres Best Pract. Res. Clin. Obstet. Gynaecol. 2000, 14, 839–854. [Google Scholar] [CrossRef] [PubMed]
- Tur-Torres, M.H.; Garrido-Gimenez, C.; Alijotas-Reig, J. Genetics of recurrent miscarriage and fetal loss. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 42, 11–25. [Google Scholar] [CrossRef]
- Practice Committee of the American Society for Reproductive Medicine. Electronic address, a.a.o. Definitions of infertility and recurrent pregnancy loss: A committee opinion. Fertil. Steril. 2020, 113, 533–535. [Google Scholar] [CrossRef]
- Ross, J.W.; Ashworth, M.D.; Stein, D.R.; Couture, O.P.; Tuggle, C.K.; Geisert, R.D. Identification of differential gene expression during porcine conceptus rapid trophoblastic elongation and attachment to uterine luminal epithelium. Physiol. Genom. 2009, 36, 140–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido-Gimenez, C.; Alijotas-Reig, J. Recurrent miscarriage: Causes, evaluation and management. Postgrad. Med. J. 2015, 91, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, O.B.; Steffensen, R.; Nielsen, H.S.; Varming, K. Multifactorial etiology of recurrent miscarriage and its scientific and clinical implications. Gynecol. Obstet. Investig. 2008, 66, 257–267. [Google Scholar] [CrossRef]
- Warren, J.E.; Silver, R.M. Genetics of pregnancy loss. Clin. Obstet. Gynecol. 2008, 51, 84–95. [Google Scholar] [CrossRef]
- Qin, W.; Tang, Y.; Yang, N.; Wei, X.; Wu, J. Potential role of circulating microRNAs as a biomarker for unexplained recurrent spontaneous abortion. Fertil. Steril. 2016, 105, 1247–1254.e3. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Li, N.; Sun, L.; Zheng, D.; Shao, G. Non-coding RNAs: The key detectors and regulators in cardiovascular disease. Genomics 2021, 113, 1233–1246. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Jain, N.; Gupta, P.; Sahoo, S.; Mallick, B. Non-coding RNAs and their cross-talks impacting reproductive health of women. Wiley Interdiscip. Rev. RNA 2021, e1695. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, P.; Zhang, L.; Huang, C.; Gao, J.; Li, Y.; Yang, B. Identification of Key Genes and Long Noncoding RNA-Associated Competing Endogenous RNA (ceRNA) Networks in Early-Onset Preeclampsia. Biomed Res. Int. 2020, 2020, 1673486. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Wang, L.; Li, S.; Wang, H.; Huang, S.; Zhang, Z.; Wang, R.; Guan, H.; Huang, Y. Identification of Key Molecules and lncRNA-miRNA-mRNA ceRNA Network in Preeclampsia. Int. J. Gen. Med. 2021, 14, 7579–7590. [Google Scholar] [CrossRef]
- Fu, X.; Cong, H.; Zhao, S.; Li, Y.; Liu, T.; Sun, Y.; Lv, N. Construction of Glycometabolism- and Hormone-Related lncRNA-Mediated Feedforward Loop Networks Reveals Global Patterns of lncRNAs and Drug Repurposing in Gestational Diabetes. Front. Endocrinol. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Zhang, C.; Ren, L.; Li, Q. Construction of a long noncoding RNA-mediated competitive endogenous RNA network reveals global patterns and regulatory markers in gestational diabetes. Int. J. Mol. Med. 2019, 43, 927–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogasawara, M.; Aoki, K.; Okada, S.; Suzumori, K. Embryonic karyotype of abortuses in relation to the number of previous miscarriages. Fertil. Steril. 2000, 73, 300–304. [Google Scholar] [CrossRef]
- Chen, X.; Guo, D.Y.; Yin, T.L.; Yang, J. Non-Coding RNAs Regulate Placental Trophoblast Function and Participate in Recurrent Abortion. Front. Pharmacol. 2021, 12, 646521. [Google Scholar] [CrossRef]
- McNamee, K.; Dawood, F.; Farquharson, R. Recurrent miscarriage and thrombophilia: An update. Curr. Opin. Obstet. Gynecol. 2012, 24, 229–234. [Google Scholar] [CrossRef]
- Kumar, S. Occupational, environmental and lifestyle factors associated with spontaneous abortion. Reprod. Sci. 2011, 18, 915–930. [Google Scholar] [CrossRef]
- Vasconcelos, S.; Ramalho, C.; Marques, C.J.; Doria, S. Altered expression of epigenetic regulators and imprinted genes in human placenta and fetal tissues from second trimester spontaneous pregnancy losses. Epigenetics 2019, 14, 1234–1244. [Google Scholar] [CrossRef]
- Nikitina, T.V.; Sazhenova, E.A.; Tolmacheva, E.N.; Sukhanova, N.N.; Kashevarova, A.A.; Skryabin, N.A.; Vasilyev, S.A.; Nemtseva, T.N.; Yuriev, S.Y.; Lebedev, I.N. Comparative Cytogenetic Analysis of Spontaneous Abortions in Recurrent and Sporadic Pregnancy Losses. Biomed. Hub 2016, 1, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sugiura-Ogasawara, M.; Ozaki, Y.; Suzumori, N. Management of recurrent miscarriage. J. Obstet. Gynaecol. Res. 2014, 40, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Pollheimer, J.; Vondra, S.; Baltayeva, J.; Beristain, A.G.; Knofler, M. Regulation of Placental Extravillous Trophoblasts by the Maternal Uterine Environment. Front. Immunol. 2018, 9, 2597. [Google Scholar] [CrossRef]
- Wu, L.; Cheng, B.; Liu, Q.; Jiang, P.; Yang, J. CRY2 suppresses trophoblast migration and invasion in recurrent spontaneous abortion. J. Biochem. 2020, 167, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tian, P.; Xu, H. MicroRNA-155-5p regulates survival of human decidua stromal cells through NF-kappaB in recurrent miscarriage. Reprod. Biol. 2021, 21, 100510. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Li, Z.; Hu, P.; Wang, J.; Fu, J.; Wei, B.; Zhang, Y. miR-219a suppresses human trophoblast cell invasion and proliferation by targeting vascular endothelial growth factor receptor 2 (VEGFR2). J. Assist. Reprod. Genet. 2021, 38, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.X.; Tang, Z.Y. Long non-coding RNA H19 regulates Bcl-2, Bax and phospholipid hydroperoxide glutathione peroxidase expression in spontaneous abortion. Exp. Ther. Med. 2021, 21, 41. [Google Scholar] [CrossRef]
- Huang, W.; Dai, M.; Qiu, T.; Liang, T.; Xie, J.; Mi, C.; Zhao, J.; Chen, W.; Tian, P.; Zhang, S.; et al. Novel lncRNA-HZ04 promotes BPDE-induced human trophoblast cell apoptosis and miscarriage by upregulating IP(3) R(1) /CaMKII/SGCB pathway by competitively binding with miR-hz04. FASEB J. 2021, 35, e21789. [Google Scholar] [CrossRef]
- Xu, Z.; Tian, P.; Guo, J.; Mi, C.; Liang, T.; Xie, J.; Huang, W.; Dai, M.; Chen, W.; Zhang, H. Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci. Total Environ. 2021, 776. [Google Scholar] [CrossRef]
- Gao, Y.; Tang, Y.; Sun, Q.; Guan, G.; Wu, X.; Shi, F.; Zhou, Z.; Yang, W. Circular RNA FOXP1 relieves trophoblastic cell dysfunction in recurrent pregnancy loss via the miR-143-3p/S100A11 cascade. Bioengineered 2021, 12, 9081–9093. [Google Scholar] [CrossRef]
- Zhu, L.; Shi, L.; Ye, W.; Li, S.; Liu, X.; Zhu, Z. Circular RNA PUM1 (CircPUM1) attenuates trophoblast cell dysfunction and inflammation in recurrent spontaneous abortion via the MicroRNA-30a-5p (miR-30a-5p)/JUNB axis. Bioengineered 2021, 12, 6878–6890. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Sayed, D.; Abdellatif, M. MicroRNAs in development and disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Elovitz, M.A.; Anton, L.; Bastek, J.; Brown, A.G. Can microRNA profiling in maternal blood identify women at risk for preterm birth? Am. J. Obstet. Gynecol. 2015, 212, 782.e1–782.e5. [Google Scholar] [CrossRef]
- Fu, G.; Brkić, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [Green Version]
- Kokawa, K.; Shikone, T.; Nakano, R. Apoptosis in human chorionic villi and decidua during normal embryonic development and spontaneous abortion in the first trimester. Placenta 1998, 19, 21–26. [Google Scholar] [CrossRef]
- Liu, H.N.; Tang, X.M.; Wang, X.Q.; Gao, J.; Li, N.; Wang, Y.Y.; Xia, H.F. MiR-93 Inhibits Trophoblast Cell Proliferation and Promotes Cell Apoptosis by Targeting BCL2L2 in Recurrent Spontaneous Abortion. Reprod. Sci. 2020, 27, 152–162. [Google Scholar] [CrossRef]
- Tian, S.; Yu, J.; Zhang, Y.; Bian, Y.; Ma, J.; Yan, J. Overexpression of PTEN regulated by miR-19b and miR-494 in the villous of recurrent spontaneous abortion patients. J. Reprod. Immunol. 2020, 140, 103133. [Google Scholar] [CrossRef] [PubMed]
- Gimm, O.; Perren, A.; Weng, L.-P.; Marsh, D.J.; Yeh, J.J.; Ziebold, U.; Gil, E.; Hinze, R.; Delbridge, L.; Lees, J.A.; et al. Differential Nuclear and Cytoplasmic Expression of PTEN in Normal Thyroid Tissue, and Benign and Malignant Epithelial Thyroid Tumors. Am. J. Pathol. 2000, 156, 1693–1700. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Shen, W.W.; Cao, X.M.; Ding, W.Y.; Yan, L.P.; Gao, L.J.; Li, X.L.; Zhong, T.Y. Novel mechanism of miRNA-365-regulated trophoblast apoptosis in recurrent miscarriage. J. Cell. Mol. Med. 2017, 21, 2412–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Lv, Y.; Wang, L.; Gong, C.; Sun, J.; Chen, X.; Chen, Y.; Yang, L.; Zhang, Y.; Yang, X.; et al. MicroRNAome in decidua: A new approach to assess the maintenance of pregnancy. Fertil. Steril. 2015, 103, 980–989.e6. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Yang, L.; Wang, H. miR-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA). Gynecol. Endocrinol. 2017, 33, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Powe, C.E.; Levine, R.J.; Karumanchi, S.A. Preeclampsia, a disease of the maternal endothelium: The role of antiangiogenic factors and implications for later cardiovascular disease. Circulation 2011, 123, 2856–2869. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Liu, J.; Fu, T.; Shan, B.; Qian, L.; Pan, L.; Yuan, J. USP25 regulates Wnt signaling by controlling the stability of tankyrases. Genes Dev. 2017, 31, 1024–1035. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Cheng, Y.; Zhang, Y.; Liao, S.; Yin, T.; Yang, J. The miR-27a-3p/USP25 axis participates in the pathogenesis of recurrent miscarriage by inhibiting trophoblast migration and invasion. J. Cell. Physiol. 2019, 234, 19951–19963. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, H.; Huo, Z.; Ma, Z.; Dang, J.; Dang, W.; Pan, L.; Chen, J.; Zhong, H. MicroRNA-16 inhibits feto-maternal angiogenesis and causes recurrent spontaneous abortion by targeting vascular endothelial growth factor. Sci. Rep. 2016, 6, 35536. [Google Scholar] [CrossRef] [Green Version]
- Chai, L.; Ling, K.; He, X.; Yang, R. Expression of ATF4 and VEGF in chorionic villus tissue in early spontaneous abortion. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 170, 434–438. [Google Scholar] [CrossRef]
- Lala, P.K. The effects of angiogenic growth factors on extravillous trophoblast invasion and motility. Placenta 2000, 21, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Girish, G.V.; Lala, N.; Di Guglielmo, G.M.; Lala, P.K. Decorin is a novel VEGFR-2-binding antagonist for the human extravillous trophoblast. Mol. Endocrinol. 2011, 25, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Chatuphonprasert, W.; Jarukamjorn, K.; Ellinger, I. Physiology and Pathophysiology of Steroid Biosynthesis, Transport and Metabolism in the Human Placenta. Front. Pharmacol. 2018, 9, 1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyall, F.; Bulmer, J.N.; Duffie, E.; Cousins, F.; Theriault, A.; Robson, S.C. Human Trophoblast Invasion and Spiral Artery Transformation. Am. J. Pathol. 2001, 158, 1713–1721. [Google Scholar] [CrossRef]
- Smith, S.D.; Dunk, C.E.; Aplin, J.D.; Harris, L.K.; Jones, R.L. Evidence for immune cell involvement in decidual spiral arteriole remodeling in early human pregnancy. Am. J. Pathol. 2009, 174, 1959–1971. [Google Scholar] [CrossRef] [Green Version]
- Wallace, A.E.; Fraser, R.; Cartwright, J.E. Extravillous trophoblast and decidual natural killer cells: A remodelling partnership. Hum. Reprod. Update 2012, 18, 458–471. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Li, J. Association of miR-34a-3p/5p, miR-141-3p/5p, and miR-24 in Decidual Natural Killer Cells with Unexplained Recurrent Spontaneous Abortion. Med. Sci. Monit. 2016, 22, 922–929. [Google Scholar] [CrossRef] [Green Version]
- He, X.; He, L.; Hannon, G.J. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007, 67, 11099–11101. [Google Scholar] [CrossRef] [Green Version]
- Fayyad-Kazan, H.; Hamade, E.; Rouas, R.; Najar, M.; Fayyad-Kazan, M.; El Zein, N.; ElDirani, R.; Hussein, N.; Fakhry, M.; Al-Akoum, C.; et al. Downregulation of microRNA-24 and -181 parallels the upregulation of IFN-γ secreted by activated human CD4 lymphocytes. Hum. Immunol. 2014, 75, 677–685. [Google Scholar] [CrossRef]
- Fiedler, J.; Jazbutyte, V.; Kirchmaier, B.C.; Gupta, S.K.; Lorenzen, J.; Hartmann, D.; Galuppo, P.; Kneitz, S.; Pena, J.T.; Sohn-Lee, C.; et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 2011, 124, 720–730. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gao, R.; Chen, X.; Zhang, H.; Zheng, A.; Yang, D.; Ding, Y.; Wang, Y.; He, J. Possible roles of mmu-miR-141 in the endometrium of mice in early pregnancy following embryo implantation. PLoS ONE 2013, 8, e67382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Li, J.; Jia, B.; Wang, Y.; Zhang, J.; Liu, G. Genome-wide identification of microRNAs in decidual natural killer cells from patients with unexplained recurrent spontaneous abortion. Am. J. Reprod. Immunol. 2018, 80, e13052. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.; Yoo, J.K.; Lee, J.; Lee, D.R.; Ko, J.J.; Oh, S.H.; Choo, Y.K.; Kim, J.K. The hsa-miR-5787 represses cellular growth by targeting eukaryotic translation initiation factor 5 (eIF5) in fibroblasts. Biochem. Biophys. Res. Commun. 2011, 415, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, H.; Zhang, Z.; Wei, R.; Zhou, X.; Wang, Z.; Zhao, L.; Guo, Q.; Zhang, Y.; Chu, C.; et al. MiR-103 protects from recurrent spontaneous abortion via inhibiting STAT1 mediated M1 macrophage polarization. Int. J. Biol. Sci. 2020, 16, 2248–2264. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhang, Y.; Cai, X.; Zhang, Y.; Yan, S.; Wang, J.; Zhang, S.; Yin, T.; Yang, C.; Yang, J. Extracellular vesicles derived from M1 macrophages deliver miR-146a-5p and miR-146b-5p to suppress trophoblast migration and invasion by targeting TRAF6 in recurrent spontaneous abortion. Theranostics 2021, 11, 5813–5830. [Google Scholar] [CrossRef]
- Cui, S.; Zhang, J.; Li, J.; Wu, H.; Zhang, H.; Yu, Q.; Zhou, Y.; Lv, X.; Zhong, Y.; Luo, S.; et al. Circulating microRNAs from serum exosomes as potential biomarkers in patients with spontaneous abortion. Am. J. Transl. Res. 2021, 13, 4197–4210. [Google Scholar]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [Green Version]
- Pan, L.; Xiao, X.; Zhao, Y.; Yin, L.; Fu, M.; Zhang, X.; Jiang, P. The functional roles of long noncoding RNA DANCR in Human Cancers. J. Cancer 2020, 11, 6970–6981. [Google Scholar] [CrossRef]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, E.L.; Belhocine, M.; Dao, L.T.; Puthier, D.; Spicuglia, S. Functions of lncRNA in development and diseases. Med. Sci. 2014, 30, 790–796. [Google Scholar] [CrossRef] [Green Version]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S. Expression of Concern: The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2016, 18, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Cao, Q.; Ge, J.; Liu, C.; Ma, Y.; Meng, Y.; Wang, Y.; Zhao, X.; Liu, R.; Li, C.; et al. LncRNA-regulated infection and inflammation pathways associated with pregnancy loss: Genome wide differential expression of lncRNAs in early spontaneous abortion. Am. J. Reprod. Immunol. 2014, 72, 359–375. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, X.; Zhu, L.; Zhu, M.; Deng, K.; Nie, X.; Mo, H.; Du, T.; Huang, B.; Hu, L.; et al. Long Noncoding RNA RP11-115N4.1 Promotes Inflammatory Responses by Interacting With HNRNPH3 and Enhancing the Transcription of HSP70 in Unexplained Recurrent Spontaneous Abortion. Front. Immunol. 2021, 12, 717785. [Google Scholar] [CrossRef]
- Wang, L.; Tang, H.; Xiong, Y.; Tang, L. Differential expression profile of long noncoding RNAs in human chorionic villi of early recurrent miscarriage. Clin. Chim. Acta 2017, 464, 17–23. [Google Scholar] [CrossRef]
- Li, T.; Li, X.; Guo, Y.; Zheng, G.; Yu, T.; Zeng, W.; Qiu, L.; He, X.; Yang, Y.; Zheng, X.; et al. Distinct mRNA and long non-coding RNA expression profiles of decidual natural killer cells in patients with early missed abortion. FASEB J. 2020, 34, 14264–14286. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Zhou, J.; Gao, Y.; Ghazal, S.; Lu, L.; Bellone, S.; Yang, Y.; Liu, N.; Zhao, X.; Santin, A.D.; et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 2015, 34, 3076–3084. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, L.; Munir, S.; Fu, G.; Dunk, C.; Baczyk, D.; Caniggia, I.; Lye, S.; Peng, C. Nodal signals through activin receptor-like kinase 7 to inhibit trophoblast migration and invasion: Implication in the pathogenesis of preeclampsia. Am. J. Pathol. 2011, 178, 1177–1189. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.J.; Cheng, Y.X.; Li, X.C.; Wang, F.; Qin, C.M.; Ma, X.L.; Yang, J.; Lin, Y. The YY1/MMP2 axis promotes trophoblast invasion at the maternal-fetal interface. J. Pathol. 2016, 239, 36–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Ding, J.; Wang, Y.; Yuan, M.; Xian, S.; Zhang, L.; Liu, S.; Dai, F.; Wang, F.; Zheng, Y.; et al. YY1-PVT1 affects trophoblast invasion and adhesion by regulating mTOR pathway-mediated autophagy. J. Cell. Physiol. 2020, 235, 6637–6646. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, F.; Li, X.C.; Shen, F.J.; Ma, X.L.; Wu, F.; Zhang, S.M.; Zeng, W.H.; Liu, X.R.; Fan, J.X.; et al. The YY1-HOTAIR-MMP2 Signaling Axis Controls Trophoblast Invasion at the Maternal-Fetal Interface. Mol. Ther. 2017, 25, 2394–2403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldassarre, A.; Masotti, A. Long non-coding RNAs and p53 regulation. Int. J. Mol. Sci. 2012, 13, 16708–16717. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.H.; Helfand, S.L. New tricks of an old molecule: Lifespan regulation by p53. Aging Cell 2006, 5, 437–440. [Google Scholar] [CrossRef] [Green Version]
- Kaare, M.; Bützow, R.; Ulander, V.M.; Kaaja, R.; Aittomäki, K.; Painter, J.N. Study of p53 gene mutations and placental expression in recurrent miscarriage cases. Reprod. Biomed. Online 2009, 18, 430–435. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.Z.; Liu, Y.; Wang, H.J.; Pang, W.W.; Zhang, J.J. Disordered p53-MALAT1 pathway is associated with recurrent miscarriage. Kaohsiung J. Med. Sci. 2019, 35, 87–94. [Google Scholar] [CrossRef]
- Wu, L.; Liu, Q.; Fan, C.; Yi, X.; Cheng, B. MALAT1 recruited the E3 ubiquitin ligase FBXW7 to induce CRY2 ubiquitin-mediated degradation and participated in trophoblast migration and invasion. J. Cell. Physiol. 2021, 236, 2169–2177. [Google Scholar] [CrossRef]
- Li, H.; Hou, Y.; Zhang, S.; Zhou, Y.; Wang, D.; Tao, S.; Ni, F. CD49a regulates the function of human decidual natural killer cells. Am. J. Reprod. Immunol. 2019, 81, e13101. [Google Scholar] [CrossRef]
- Sheng, F.; Sun, N.; Ji, Y.; Ma, Y.; Ding, H.; Zhang, Q.; Yang, F.; Li, W. Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion. J. Cell. Biochem. 2019, 120, 17378–17390. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Du, G.; Huang, X.; Han, L.; Han, X.; Xu, B.; Zhang, Y.; Yu, M.; Qin, Y.; Xia, Y.; et al. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine 2018, 38, 162–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, D.; Zeng, H.; Chen, J.; Xiao, L.; Zhao, Y.; Liu, N. H19 regulates trophoblastic spheroid adhesion by competitively binding to let-7. Reproduction 2019, 157, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Yan, H.; Sun, B.; Feng, F.; Chen, P. Decreased expression of long non-coding RNA SNHG7 cause recurrent spontaneous abortion through suppression proliferation and invasion of trophoblast cells via miR-34a. Am. J. Transl. Res. 2019, 11, 463–472. [Google Scholar]
- Li, Z.; Zhou, G.; Tao, F.; Cao, Y.; Han, W.; Li, Q. circ-ZUFSP regulates trophoblasts migration and invasion through sponging miR-203 to regulate STOX1 expression. Biochem. Biophys. Res. Commun. 2020, 531, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Huang, C.; Wang, X.; Shan, G. Circular RNAs in Eukaryotic Cells. Curr. Genomics 2015, 16, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Hanan, M.; Soreq, H.; Kadener, S. CircRNAs in the brain. RNA Biol. 2017, 14, 1028–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.H.; Xiao, Z.; Tong, J.H.; To, K.F.; Fang, X.; Cheng, A.S.; Chen, Y. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int. J. Cancer 2017, 140, 120–129. [Google Scholar] [CrossRef]
- Du, W.W.; Zhang, C.; Yang, W.; Yong, T.; Awan, F.M.; Yang, B.B. Identifying and Characterizing circRNA-Protein Interaction. Theranostics 2017, 7, 4183–4191. [Google Scholar] [CrossRef]
- Dong, W.; Dai, Z.H.; Liu, F.C.; Guo, X.G.; Ge, C.M.; Ding, J.; Liu, H.; Yang, F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 2019, 45, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef] [Green Version]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W.; Dou, K.; Li, H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, I.; Karedath, T.; Andrews, S.S.; Al-Azwani, I.K.; Mohamoud, Y.A.; Querleu, D.; Rafii, A.; Malek, J.A. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 2016, 7, 36366–36381. [Google Scholar] [CrossRef] [Green Version]
- Xin, Z.; Ma, Q.; Ren, S.; Wang, G.; Li, F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct. Genom. 2017, 16, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, X.; Liu, X.; Liu, X.; He, J.; Ding, Y.; Tong, C.; Peng, C.; Wang, Y.; Gao, R. CircRNA expression profiles in decidual tissue of patients with early recurrent miscarriage. Genes Dis. 2020, 7, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Wang, X.; Ruan, H.; Rui, C.; Mao, P.; Cheng, Q.; Jia, R. Circular RNAs expressed in chorionic villi are probably involved in the occurrence of recurrent spontaneous abortion. Biomed. Pharmacother. 2017, 88, 1154–1162. [Google Scholar] [CrossRef]
- Wu, X.; Sui, Z.; Zhang, H.; Wang, Y.; Yu, Z. Integrated Analysis of lncRNA-Mediated ceRNA Network in Lung Adenocarcinoma. Front. Oncol. 2020, 10, 554759. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.S.; Zhang, E.X.; Sun, Q.F.; Ye, Z.J.; Liu, J.W.; Zhou, D.H.; Tang, Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 2019, 19, 779. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Bai, Y.; Yang, X.; Lin, J.; Yang, X.; Wang, D.; He, L.; Zheng, Y.; Zhao, H. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019, 19, 90. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, M.; Cao, Y.; Zhang, D.; Han, M.; Gao, X.; Xu, B.; Zhang, A. Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene 2019, 720, 144056. [Google Scholar] [CrossRef]
- Huang, Y.; Hao, J.; Liao, Y.; Zhou, L.; Wang, K.; Zou, H.; Hu, Y.; Li, J. Transcriptome sequencing identified the ceRNA network associated with recurrent spontaneous abortion. BMC Med. Genomics 2021, 14, 278. [Google Scholar] [CrossRef]
- Zang, X.; Gu, T.; Wang, W.; Zhou, C.; Ding, Y.; Gu, S.; Xu, Z.; Xie, Y.; Li, Z.; Cai, G.; et al. Integrated Insight into the Molecular Mechanisms of Spontaneous Abortion during Early Pregnancy in Pigs. Int. J. Mol. Sci. 2021, 22, 6644. [Google Scholar] [CrossRef]
- Wang, M.M.; Zhong, J.X.; Xiang, Y.Y. LncRNA-GAS5 related to the processes of recurrent pregnancy loss by regulating Th1/Th2 balance. Kaohsiung J. Med. Sci. 2021, 37, 479–486. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Li, J.; Mao, B. Urothelial cancer associated 1 (UCA1) regulates trophoblast viability, proliferation, and migration via modulating the UCA1/miR-455/RUNX2 signaling pathway. Acta Biochim. Biophys. Sin. 2020, 52, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, T.B.; Veno, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Li, Y.; Liu, W.; He, J.; Zhang, L.; Li, H.; Li, P.; Lv, L. Comprehensive circRNA expression profile and construction of circRNA-associated ceRNA network in fur skin. Exp. Dermatol. 2018, 27, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Gu, T.; He, Y.; Zhou, C.; Hu, Q.; Wang, X.; Zheng, E.; Huang, S.; Xu, Z.; Yang, J.; et al. Genome-Wide Analysis of Circular RNAs Mediated ceRNA Regulation in Porcine Embryonic Muscle Development. Front. Cell Dev. Biol. 2019, 7, 289. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Chen, L.; Shan, Y.; Chen, G.; Chu, Y.; Dai, H.; Liu, X.; Bao, H. Hsa_circ_0038383-mediated competitive endogenous RNA network in recurrent implantation failure. Aging 2021, 13, 6076–6090. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhao, H.; Gong, L.; Xiao, X.; Zhou, Q.; Lu, H.; Cui, Y.; Xu, H.; Wu, S.; Tang, Y.; et al. Differentially expressed circular RNAs and the competing endogenous RNA network associated with preeclampsia. Placenta 2021, 103, 232–241. [Google Scholar] [CrossRef]
- Jin, D.; Wu, X.; Yu, H.; Jiang, L.; Zhou, P.; Yao, X.; Meng, J.; Wang, L.; Zhang, M.; Zhang, Y. Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am. J. Transl. Res. 2018, 10, 1498–1510. [Google Scholar]
- Chen, D.; He, B.; Zheng, P.; Wang, S.; Zhao, X.; Liu, J.; Yang, X.; Cheng, W. Identification of mRNA-, circRNA- and lncRNA- Associated ceRNA Networks and Potential Biomarkers for Preeclampsia From Umbilical Vein Endothelial Cells. Front. Mol. Biosci. 2021, 8, 652250. [Google Scholar] [CrossRef]
- Prather, R.S.; Lorson, M.; Ross, J.W.; Whyte, J.J.; Walters, E. Genetically engineered pig models for human diseases. Annu Rev Anim. Biosci. 2013, 1, 203–219. [Google Scholar] [CrossRef] [Green Version]
- Jia, N.; Li, J. Noncoding RNAs in Unexplained Recurrent Spontaneous Abortions and Their Diagnostic Potential. Dis. Markers 2019, 2019, 7090767. [Google Scholar] [CrossRef] [Green Version]
- Viereck, J.; Thum, T. Circulating Noncoding RNAs as Biomarkers of Cardiovascular Disease and Injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef] [PubMed]


| miRNA Names | Expression in SA | Target | Tissue | Function | Reference |
|---|---|---|---|---|---|
| MiR-93 | Upregulation | BCL2L2 | Chorionic villi, trophoblast | Promotes proliferation, migration, invasion, and apoptosis | [40] |
| MiR-19b | Upregulation | PTEN | Placental villi | Regulates abnormal cellular invasion and apoptosis | [41] |
| MiR-494 | Downregulation | PTEN | Placental villi | Regulates abnormal cellular invasion and apoptosis | [41] |
| MiR-365 | Upregulation | SGK1 | Decidua | Regulates cellular apoptosis | [43] |
| MiR-199B-5p | Upregulation | SGK1 | Decidua | Regulates cellular apoptosis | [44] |
| MiR-520 | Upregulation | PARP1 | Trophoblast | Promotes DNA damage-induced apoptosis | [45] |
| MiR-27A-3p | Upregulation | USP25 | Trophoblast | Regulates the process of EMT, cell invasion, and migration | [48] |
| MiR-16 | Upregulation | VEGF | Placenta | Regulates angiogenesis and development | [49] |
| MiR-219a | Downregulation | VEGFR2 | Trophoblast | Negative regulation of cellular proliferation and invasion | [26] |
| MiR-155-5p | Downregulation | NF-kB pathway | Decidual stroma | Promotes cellular growth and proliferation and inhibits apoptosis | [25] |
| MiR-34a-3p/5p | Upregulation | P53 | NK cells | Regulates cellular apoptosis and proliferation | [57,58] |
| MiR-141-3p/5p | Upregulation | PTEN | NK cells | Regulates cellular apoptosis and proliferation | [57,61] |
| MiR-24 | Downregulation | IFNG | NK cells | Regulates cellular apoptosis and proliferation | [57,59,60] |
| Hsa-miR-5787 | Upregulation | EIF5 | NK cells | Inhibits cellular growth | [62,63] |
| MiR-103 | Downregulation | STAT1/IRF1 pathway | M1 macrophages | Negative regulation of cellular polarization | [64] |
| MiR-146a/b-5p | Upregulation | TRAF6 | M1 macrophages | Inhibits EMT and maintains cellular interaction | [65] |
| RNA Names | Expression in SA | Target | Tissue | Function | Reference |
|---|---|---|---|---|---|
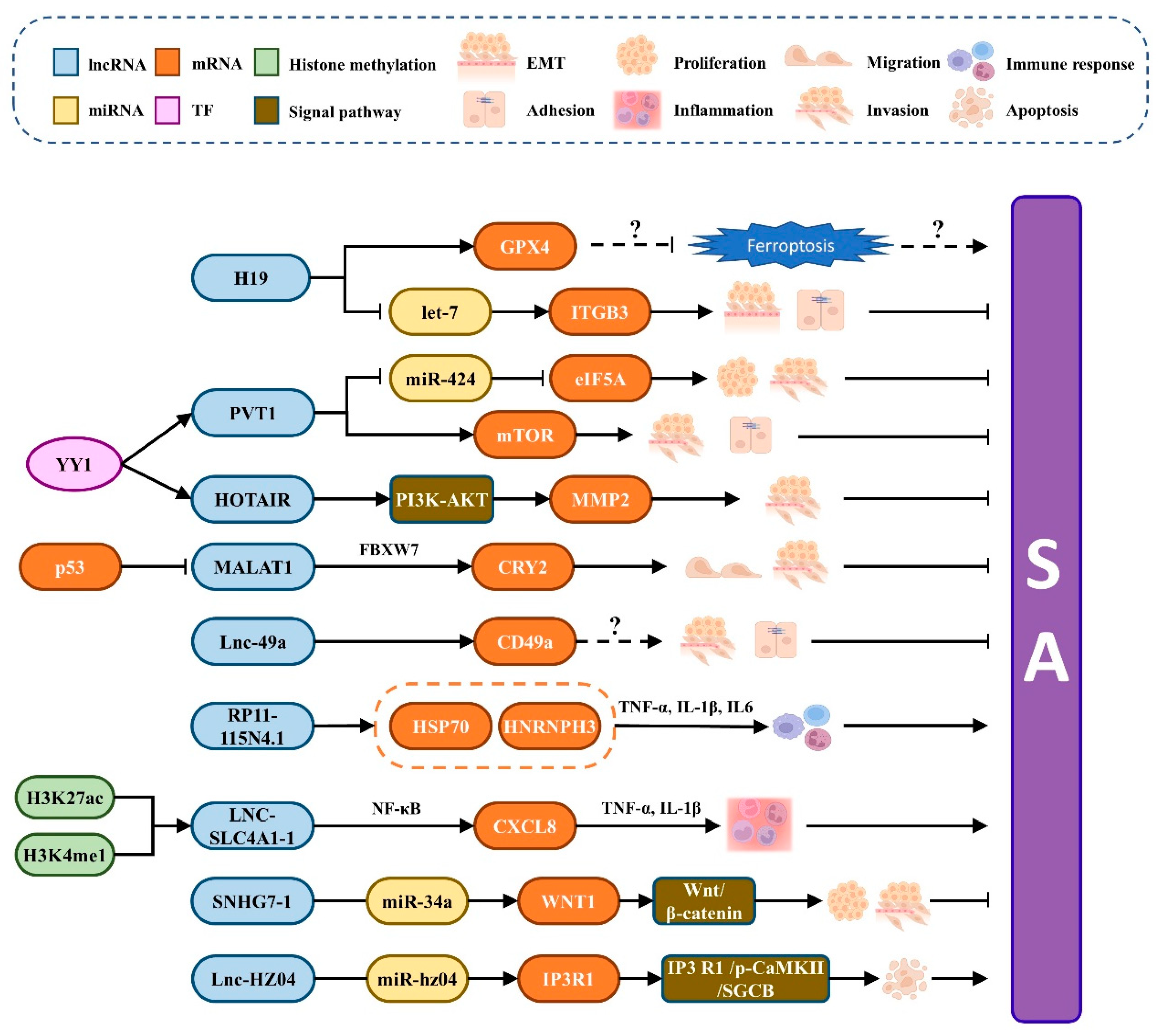
| H19 | Downregulation | GPX4 | Placental villi | Regulates invasion and migration, and may be related to ferroptosis. | [80] |
| Let-7 | Competes with miRNA let-7 to control the target gene ITGB3 | [93,94] | |||
| PVT1 | Downregulation | MTOR | Trophoblast | Control apoptosis, adhesion, and invasion through the mTOR pathway. | [82,83] |
| MiR-424 | Regulate the function of trophoblast cells through the PVT1/miR-424/eIF5A pathway | [83] | |||
| HOTAIR | Downregulation | PI3K-AKT | Trophoblast | Promotes the production of matrix metalloproteinase 2 and enhances cell invasion | [84,85] |
| MALAT1 | Downregulation | FBXW7 | Trophoblast | Inhibits SA by playing a key role in the P53-MALAT1-FBXW7-CRY2 axis. | [88,89] |
| Lnc-49a | Downregulation | CD49a | DNK cells | Impedes SA by regulating the expression of CD49a | [90] |
| RP11-115N4.1 | Upregulation | PARP1 | Peripheral blood | Inhibits the migration of trophoblast cells by increasing HSP70 and HNRNPH3. | [74] |
| MEG8 | Upregulation | Unknown | Chorion villi | Inhibits cell proliferation and invasion in trophoblast cell lines | [91] |
| LNC-SLC4A1-1 | Upregulation | CXCL8 | URSA Villi | Intensifies trophoblast cell inflammation by activating CXCL8 | [92] |
| SNHG7-1 | Upregulation | MiR-34a | Placental villi | Promotes the occur-rence of RSA via the Wnt/β-catenin signaling pathway | [95] |
| Lnc-HZ04 | Upregulation | MiR-hz04 | Villi | Weakens the decreasing effect of miR-hz04 on inositol IP3R1 expression and stability | [28] |
| CIRC-ZUFSP | Upregulation | MiR-203 | Trophoblast | Inhibits migration and invasion of trophoblast cells through the CIRC-ZUFSP/miR-203/STOX1 pathway | [96] |
| CIRCPUM1 | Downregulation | MiR-30a-5p | Placenta | Promotes trophoblast cell processes and anti-inflammatory effects through the miR-30a-5p/JunB axis | [31] |
| CircFOXP1 | Downregulation | MiR-143-3p | Trophoblast | Regulates the function of trophoblast cells via the miR-143-3p/S100A11 pathway | [30] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, J.; Gu, T.; Yang, H.; Ao, Z.; Cai, G.; Hong, L.; Wu, Z. Non-Coding RNAs Regulate Spontaneous Abortion: A Global Network and System Perspective. Int. J. Mol. Sci. 2022, 23, 4214. https://doi.org/10.3390/ijms23084214
Gan J, Gu T, Yang H, Ao Z, Cai G, Hong L, Wu Z. Non-Coding RNAs Regulate Spontaneous Abortion: A Global Network and System Perspective. International Journal of Molecular Sciences. 2022; 23(8):4214. https://doi.org/10.3390/ijms23084214
Chicago/Turabian StyleGan, Jianyu, Ting Gu, Huaqiang Yang, Zheng Ao, Gengyuan Cai, Linjun Hong, and Zhenfang Wu. 2022. "Non-Coding RNAs Regulate Spontaneous Abortion: A Global Network and System Perspective" International Journal of Molecular Sciences 23, no. 8: 4214. https://doi.org/10.3390/ijms23084214






