Recognition of a Novel Gene Signature for Human Glioblastoma
Abstract
:1. Introduction
2. Results
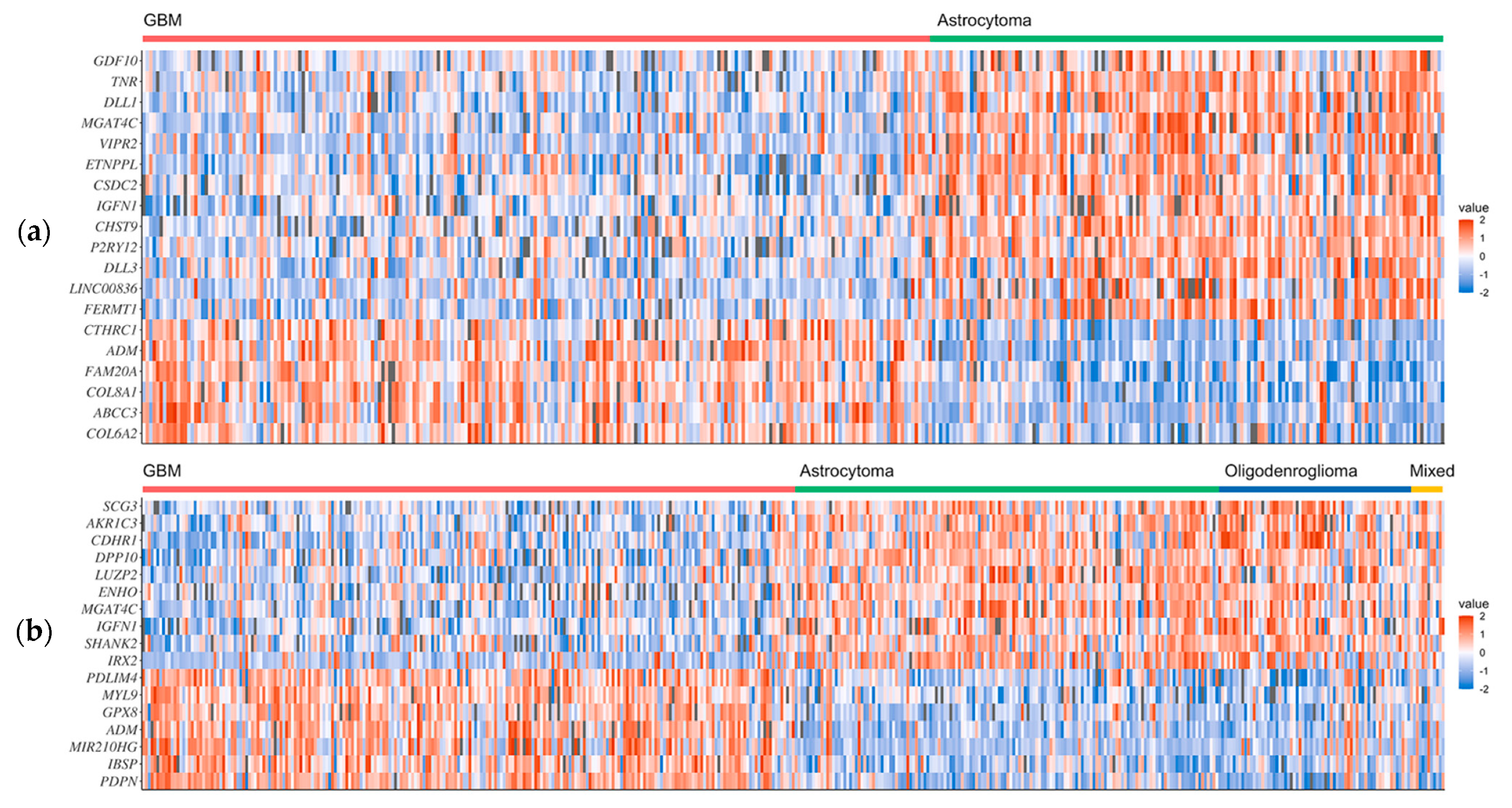
2.1. Recognition of Novel Gene Signatures in GBM
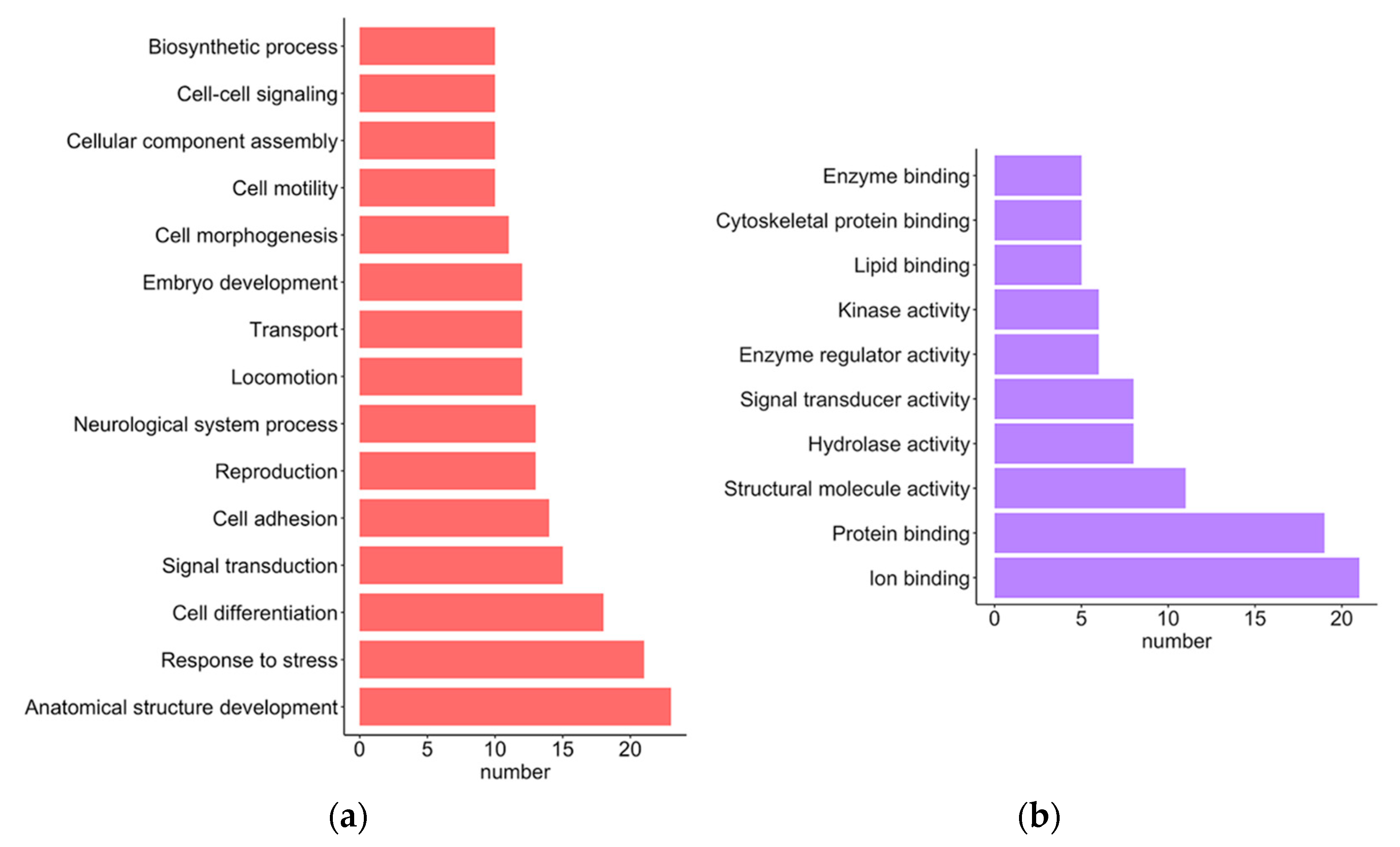
2.2. Functional Analysis
2.3. Roles of the Overexpressed GBM Gene Signatures
2.4. Roles of the Underexpressed GBM Gene Signatures
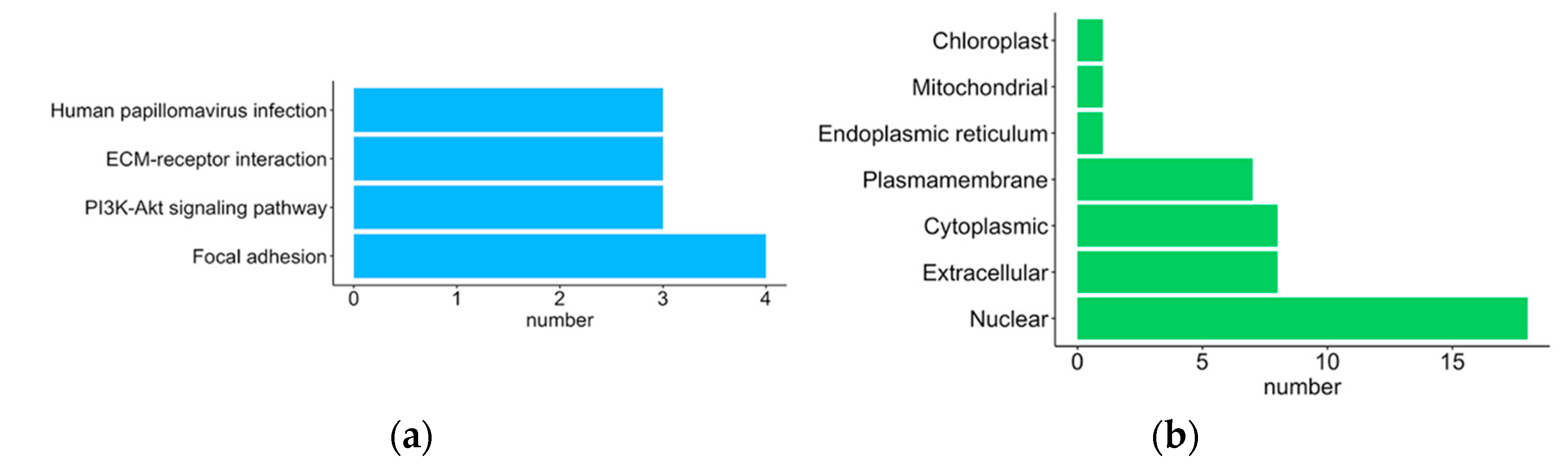
2.5. Protein–Protein Interaction Network Analysis Using STRING
3. Discussion
4. Materials and Methods
4.1. Brain Tumor Dataset
4.2. Gene Feature Generation
4.3. Selection of Critical Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrom, Q.T.; Gittleman, H.; Farah, P.; Ondracek, A.; Chen, Y.; Wolinsky, Y.; Stroup, N.E.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013, 15 (Suppl. 2), ii1–ii56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Klockow, J.L.; Zhang, M.; Lafortune, F.; Chang, E.; Jin, L.; Wu, Y.; Daldrup-Link, H.E. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharm. Res. 2021, 171, 105780. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, V.E.; Solheim, O.; Salvesen, O.; Torp, S.H. The histological representativeness of glioblastoma tissue samples. Acta Neurochir. 2021, 163, 1911–1920. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velazquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef] [Green Version]
- Hara, A.; Kanayama, T.; Noguchi, K.; Niwa, A.; Miyai, M.; Kawaguchi, M.; Ishida, K.; Hatano, Y.; Niwa, M.; Tomita, H. Treatment Strategies Based on Histological Targets against Invasive and Resistant Glioblastoma. J. Oncol. 2019, 2019, 2964783. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Ma, X.; Deng, X.; Ji, T.; Hu, P.; Wan, R.; Qiu, H.; Cui, D.; Gao, L. Identification of key candidate genes and pathways in glioblastoma by integrated bioinformatical analysis. Exp. Med. 2019, 18, 3439–3449. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Shi, Y.; Zhao, W.; Zhang, Y.; Xie, Y.; Zhang, B.; Tan, G.; Wang, Z. Development of an Immune-Related Prognostic Index Associated with Glioblastoma. Front. Neurol. 2021, 12, 610797. [Google Scholar] [CrossRef]
- Yin, W.; Tang, G.; Zhou, Q.; Cao, Y.; Li, H.; Fu, X.; Wu, Z.; Jiang, X. Expression Profile Analysis Identifies a Novel Five-Gene Signature to Improve Prognosis Prediction of Glioblastoma. Front. Genet. 2019, 10, 419. [Google Scholar] [CrossRef] [PubMed]
- Zottel, A.; Jovcevska, I.; Samec, N.; Komel, R. Cytoskeletal proteins as glioblastoma biomarkers and targets for therapy: A systematic review. Crit. Rev. Oncol. Hematol. 2021, 160, 103283. [Google Scholar] [CrossRef] [PubMed]
- Kruthika, B.S.; Sugur, H.; Nandaki, K.; Arimappamagan, A.; Paturu, K.; Santosh, V. Expression pattern and prognostic significance of myosin light chain 9 (MYL9): A novel biomarker in glioblastoma. J. Clin. Pathol. 2019, 72, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wang, P.; Ning, S.; Xiao, W.; Xiao, B.; Li, X. Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget 2016, 7, 41737–41747. [Google Scholar] [CrossRef] [Green Version]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Saadeh, F.S.; Mahfouz, R.; Assi, H.I. EGFR as a clinical marker in glioblastomas and other gliomas. Int. J. Biol. Mark. 2018, 33, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Loureiro, L.V.M.; Neder, L.; Callegaro-Filho, D.; de Oliveira Koch, L.; Stavale, J.N.; Malheiros, S.M.F. The immunohistochemical landscape of the VEGF family and its receptors in glioblastomas. Surg. Exp. Pathol. 2020, 3, 9. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, S.; Zhang, M.; Yang, L.; Zhong, J.; Li, B.; Li, F.; Xia, X.; Li, X.; Zhou, H.; et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021, 22, 33. [Google Scholar] [CrossRef]
- Gu, W.; Shou, J.; Gu, S.; Sun, B.; Che, X. Identifying hedgehog signaling specific microRNAs in glioblastomas. Int. J. Med. Sci. 2014, 11, 488–493. [Google Scholar] [CrossRef] [Green Version]
- Bazzoni, R.; Bentivegna, A. Role of Notch Signaling Pathway in Glioblastoma Pathogenesis. Cancers 2019, 11, 292. [Google Scholar] [CrossRef] [Green Version]
- Latour, M.; Her, N.G.; Kesari, S.; Nurmemmedov, E. WNT Signaling as a Therapeutic Target for Glioblastoma. Int. J. Mol. Sci. 2021, 22, 8428. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fang, R.; Yue, C.; Chang, G.; Li, P.; Guo, Q.; Wang, J.; Zhou, A.; Zhang, S.; Fuller, G.N.; et al. Wnt-Induced Stabilization of KDM4C Is Required for Wnt/beta-Catenin Target Gene Expression and Glioblastoma Tumorigenesis. Cancer Res. 2020, 80, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Sasmita, A.O.; Wong, Y.P.; Ling, A.P.K. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac. J. Clin. Oncol. 2018, 14, 40–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, X.; Xu, L.; Lin, Y.; Zhang, J.; Cao, H. Low expression of CDHR1 is an independent unfavorable prognostic factor in glioma. J. Cancer 2021, 12, 5193–5205. [Google Scholar] [CrossRef]
- Talukdar, S.; Das, S.K.; Pradhan, A.K.; Emdad, L.; Shen, X.N.; Windle, J.J.; Sarkar, D.; Fisher, P.B. Novel function of MDA-9/Syntenin (SDCBP) as a regulator of survival and stemness in glioma stem cells. Oncotarget 2016, 7, 54102–54119. [Google Scholar] [CrossRef] [Green Version]
- Maimaiti, A.; Wang, X.; Hao, Y.; Jiang, L.; Shi, X.; Pei, Y.; Feng, Z.; Kasimu, M. Integrated Gene Expression and Methylation Analyses Identify DLL3 as a Biomarker for Prognosis of Malignant Glioma. J. Mol. Neurosci. 2021, 71, 1622–1635. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, N.; Wang, J.; Cao, J.; Li, D.; Zhang, Y.; Zhang, L. SCG3 Protein Expression in Glioma Associates With less Malignancy and Favorable Clinical Outcomes. Pathol. Oncol. Res. 2021, 27, 594931. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef]
- Thai, M.T.; Wu, W.; Xiong, H. Big Data in Complex and Social Networks, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2016. [Google Scholar] [CrossRef]
- Gusev, Y.; Bhuvaneshwar, K.; Song, L.; Zenklusen, J.C.; Fine, H.; Madhavan, S. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data 2018, 5, 180158. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. PLoS ONE 2014, 9, e99368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Tayrac, M.; Saikali, S.; Aubry, M.; Bellaud, P.; Boniface, R.; Quillien, V.; Mosser, J. Prognostic significance of EDN/RB, HJURP, p60/CAF-1 and PDLI4, four new markers in high-grade gliomas. PLoS ONE 2013, 8, e73332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, C.; He, K.; Li, S.; Ge, Q.; Liu, L.; Zhang, Z.; Zhang, H.; Wang, X.; Sun, X.; Ding, L. ITPRIP promotes glioma progression by linking MYL9 to DAPK1 inhibition. Cell Signal. 2021, 85, 110062. [Google Scholar] [CrossRef]
- Sun, Z.; Qi, X.; Zhang, Y. Bioinformatics Analysis of the Expression of ATP Binding Cassette Subfamily C Member 3 (ABCC3) in Human Glioma. Open Med. 2020, 15, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.Y.; Li, X.D.; Ma, W.N.; Li, H.; Li, M.M.; Yang, X.Y.; Li, S.Y. Comprehensive Transcriptomic Analysis and Experimental Validation Identify lncRNA HOXA-AS2/miR-184/COL6A2 as the Critical ceRNA Regulation Involved in Low-Grade Glioma Recurrence. Onco Targets 2020, 13, 4999–5016. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, J.; Zhao, L.; Wang, X.; Ma, D.; Xu, B.; Xie, F.; Qi, X.; Chen, G.; Zhao, H.; et al. Fam20C Overexpression Predicts Poor Outcomes and is a Diagnostic Biomarker in Lower-Grade Glioma. Front. Genet. 2021, 12, 757014. [Google Scholar] [CrossRef]
- Lim, S.Y.; Ahn, S.H.; Park, H.; Lee, J.; Choi, K.; Choi, C.; Choi, J.H.; Park, E.M.; Choi, Y.H. Transcriptional regulation of adrenomedullin by oncostatin M in human astroglioma cells: Implications for tumor invasion and migration. Sci. Rep. 2014, 4, 6444. [Google Scholar] [CrossRef] [Green Version]
- Mei, P.J.; Bai, J.; Miao, F.A.; Chen, C.; Zhu, Y.S.; Li, Z.L.; Zheng, J.N.; Fan, Y.C. CTHRC1 mediates multiple pathways regulating cell invasion, migration and adhesion in glioma. Int. J. Clin. Exp. Pathol. 2017, 10, 9318–9329. [Google Scholar] [PubMed]
- Du, S.; Guan, S.; Zhu, C.; Guo, Q.; Cao, J.; Guan, G.; Cheng, W.; Cheng, P.; Wu, A. Secretory Pathway Kinase FAM20C, a Marker for Glioma Invasion and Malignancy, Predicts Poor Prognosis of Glioma. Onco Targets 2020, 13, 11755–11768. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Luo, H.; Chen, L.; Li, J.; Zhu, X.; Huang, K. Identification of an epithelial-mesenchymal transition related long non-coding RNA (LncRNA) signature in Glioma. Bioengineered 2021, 12, 4016–4031. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Dai, D.; Wang, J.; Zhang, D.; Zhang, Y.; Han, G.; Zhang, L.; Chen, C.; Li, X.; Li, Y.; et al. Long Noncoding RNA miR210HG as a Potential Biomarker for the Diagnosis of Glioma. PLoS ONE 2016, 11, e0160451. [Google Scholar] [CrossRef] [Green Version]
- Grau, S.J.; Trillsch, F.; Tonn, J.C.; Goldbrunner, R.H.; Noessner, E.; Nelson, P.J.; von Luettichau, I. Podoplanin increases migration and angiogenesis in malignant glioma. Int. J. Clin. Exp. Pathol. 2015, 8, 8663–8670. [Google Scholar]
- Serao, N.V.; Delfino, K.R.; Southey, B.R.; Beever, J.E.; Rodriguez-Zas, S.L. Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC Med. Genom. 2011, 4, 49. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Zhao, X.; Cui, L.; He, G.; Wang, X.; Wang, F.; Duan, S.; He, L.; Li, Q.; Yu, X.; et al. Genetic regulatory subnetworks and key regulating genes in rat hippocampus perturbed by prenatal malnutrition: Implications for major brain disorders. Aging 2020, 12, 8434–8458. [Google Scholar] [CrossRef]
- Bi, B.; Li, F.; Guo, J.; Li, C.; Jing, R.; Lv, X.; Chen, X.; Wang, F.; Azadzoi, K.M.; Wang, L.; et al. Label-free quantitative proteomics unravels the importance of RNA processing in glioma malignancy. Neuroscience 2017, 351, 84–95. [Google Scholar] [CrossRef]
- Park, A.L.; Lin, H.K.; Yang, Q.; Sing, C.W.; Fan, M.; Mapstone, T.B.; Gross, N.L.; Gumerlock, M.K.; Martin, M.D.; Rabb, C.H.; et al. Differential expression of type 2 3alpha/type 5 17beta-hydroxysteroid dehydrogenase (AKR1C3) in tumors of the central nervous system. Int. J. Clin. Exp. Pathol. 2010, 3, 743–754. [Google Scholar]
- Gonzalez-Garcia, N.; Nieto-Librero, A.B.; Vital, A.L.; Tao, H.J.; Gonzalez-Tablas, M.; Otero, A.; Galindo-Villardon, P.; Orfao, A.; Tabernero, M.D. Multivariate analysis reveals differentially expressed genes among distinct subtypes of diffuse astrocytic gliomas: Diagnostic implications. Sci. Rep. 2020, 10, 11270. [Google Scholar] [CrossRef]
- Antonelli, M.; Fadda, A.; Loi, E.; Moi, L.; Zavattari, C.; Sulas, P.; Gentilini, D.; Cameli, C.; Bacchelli, E.; Badiali, M.; et al. Integrated DNA methylation analysis identifies topographical and tumoral biomarkers in pilocytic astrocytomas. Oncotarget 2018, 9, 13807–13821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Kros, J.M.; van der Weiden, M.; Zheng, P.; Cheng, C.; Mustafa, D.A. Expression site of P2RY12 in residential microglial cells in astrocytomas correlates with M1 and M2 marker expression and tumor grade. Acta Neuropathol. Commun. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaworski, D.M. Expression of pituitary adenylate cyclase-activating polypeptide (PACAP) and the PACAP-selective receptor in cultured rat astrocytes, human brain tumors, and in response to acute intracranial injury. Cell Tissue Res. 2000, 300, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, L.; Shan, Z.L.; Chen, S.; Hu, H. GPX8 is transcriptionally regulated by FOXC1 and promotes the growth of gastric cancer cells through activating the Wnt signaling pathway. Cancer Cell Int. 2020, 20, 596. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Guo, Y.; Zhao, Q. GPX8 promotes migration and invasion by regulating epithelial characteristics in non-small cell lung cancer. Thorac. Cancer 2020, 11, 3299–3308. [Google Scholar] [CrossRef]
- Khatib, A.; Solaimuthu, B.; Ben Yosef, M.; Abu Rmaileh, A.; Tanna, M.; Oren, G.; Schlesinger Frisch, M.; Axelrod, J.H.; Lichtenstein, M.; Shaul, Y.D. The glutathione peroxidase 8 (GPX8)/IL-6/STAT3 axis is essential in maintaining an aggressive breast cancer phenotype. Proc. Natl. Acad. Sci USA 2020, 117, 21420–21431. [Google Scholar] [CrossRef]
- Shahcheraghi, S.H.; Tchokonte-Nana, V.; Lotfi, M.; Lotfi, M.; Ghorbani, A.; Sadeghnia, H.R. Wnt/beta-catenin and PI3K/Akt/mTOR Signaling Pathways in Glioblastoma: Two Main Targets for Drug Design: A Review. Curr. Pharm. Des. 2020, 26, 1729–1741. [Google Scholar] [CrossRef]
- Guan, R.; Zhang, X.; Guo, M. Glioblastoma stem cells and Wnt signaling pathway: Molecular mechanisms and therapeutic targets. Chin. Neurosurg. J. 2020, 6, 25. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, X.; Shi, Y.; Zhang, M.; Yang, J.; Dong, M.; Mi, Y.; Zhang, Z.; Liu, K.; Jiang, L.; et al. FOXC1 silencing inhibits the epithelialtomesenchymal transition of glioma cells: Involvement of betacatenin signaling. Mol. Med. Rep. 2019, 19, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Kim, B.H.; Lee, H.; Park, C.G.; Jeong, A.J.; Lee, S.H.; Noh, K.H.; Park, J.B.; Lee, C.G.; Paek, S.H.; Kim, H.; et al. STAT3 Inhibitor ODZ10117 Suppresses Glioblastoma Malignancy and Prolongs Survival in a Glioblastoma Xenograft Model. Cells 2020, 9, 722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Zhang, N.; Zhu, H.; Liu, J.; Xing, H.; Ma, F.; Yang, M. CHST9 rs1436904 genetic variant contributes to prognosis of triple-negative breast cancer. Sci. Rep. 2017, 7, 11802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Wu, Q.; Fu, X.; Yu, B.; Shao, Y.; Yang, H.; Guan, M.; Huang, X.; Zhang, W.; Wan, J. Examination of copy number variations of CHST9 in multiple types of hematologic malignancies. Cancer Genet. Cytogenet. 2010, 203, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Su, Z.; Yang, B.; Zeng, Z.; Lei, S.; Qiao, H. Identification of key genes involved in the development and progression of early-onset colorectal cancer by co-expression network analysis. Oncol. Lett. 2020, 19, 177–186. [Google Scholar] [CrossRef] [Green Version]
- Xing, Q.; Liu, S.; Luan, J.; Wang, Y.; Ma, L. A novel 13 RNA binding proteins (RBPs) signature could predict prostate cancer biochemical recurrence. Pathol. Res. Pr. 2021, 225, 153587. [Google Scholar] [CrossRef] [PubMed]
- Jasaszwili, M.; Billert, M.; Strowski, M.Z.; Nowak, K.W.; Skrzypski, M. Adropin as A Fat-Burning Hormone with Multiple Functions-Review of a Decade of Research. Molecules 2020, 25, 549. [Google Scholar] [CrossRef] [Green Version]
- Nergiz, S.; Altinkaya, S.O.; Kurt Omurlu, I.; Yuksel, H.; Kucuk, M.; Demircan Sezer, S. Circulating adropin levels in patients with endometrium cancer. Gynecol. Endocrinol. 2015, 31, 730–735. [Google Scholar] [CrossRef]
- Wang, X.; Chen, Q. FERMT1 knockdown inhibits oral squamous cell carcinoma cell epithelial-mesenchymal transition by inactivating the PI3K/AKT signaling pathway. BMC Oral Health 2021, 21, 598. [Google Scholar] [CrossRef]
- Miao, C.; Liu, D.; Zhang, F.; Wang, Y.; Zhang, Y.; Yu, J.; Zhang, Z.; Liu, G.; Li, B.; Liu, X.; et al. Association of FPGS genetic polymorphisms with primary retroperitoneal liposarcoma. Sci. Rep. 2015, 5, 9079. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.P.; Das, P. Novel splicing in IGFN1 intron 15 and role of stable G-quadruplex in the regulation of splicing in renal cell carcinoma. PLoS ONE 2018, 13, e0205660. [Google Scholar] [CrossRef]
- Ma, Q.; Geng, K.; Xiao, P.; Zeng, L. Identification and Prognostic Value Exploration of Radiotherapy Sensitivity-Associated Genes in Non-Small-Cell Lung Cancer. Biomed. Res. Int. 2021, 2021, 5963868. [Google Scholar] [CrossRef] [PubMed]
- Demichelis, F.; Setlur, S.R.; Banerjee, S.; Chakravarty, D.; Chen, J.Y.; Chen, C.X.; Huang, J.; Beltran, H.; Oldridge, D.A.; Kitabayashi, N.; et al. Identification of functionally active, low frequency copy number variants at 15q21.3 and 12q21.31 associated with prostate cancer risk. Proc. Natl. Acad. Sci. USA 2012, 109, 6686–6691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Li, P.; Hao, X.; Lu, Y.; Liu, M.; Song, W.; Shan, L.; Yu, J.; Ding, H.; Chen, S.; et al. SHANK2 is a frequently amplified oncogene with evolutionarily conserved roles in regulating Hippo signaling. Protein Cell 2021, 12, 174–193. [Google Scholar] [CrossRef]
- Masliantsev, K.; Karayan-Tapon, L.; Guichet, P.O. Hippo Signaling Pathway in Gliomas. Cells 2021, 10, 184. [Google Scholar] [CrossRef]
- Xue, J.; Sang, W.; Su, L.P.; Gao, H.X.; Cui, W.L.; Abulajiang, G.; Wang, Q.; Zhang, J.; Zhang, W. Proteomics reveals protein phosphatase 1gamma as a biomarker associated with Hippo signal pathway in glioma. Pathol. Res. Pr. 2020, 216, 153187. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, P.; Lundgren, I.; Mukohyama, H.; Lehenkari, P.P.; Horton, M.A.; Lerner, U.H. Vasoactive intestinal peptide (VIP)/pituitary adenylate cyclase-activating peptide receptor subtypes in mouse calvarial osteoblasts: Presence of VIP-2 receptors and differentiation-induced expression of VIP-1 receptors. Endocrinology 2001, 142, 339–347. [Google Scholar] [CrossRef]
- D’Amico, A.G.; Scuderi, S.; Saccone, S.; Castorina, A.; Drago, F.; D’Agata, V. Antiproliferative effects of PACAP and VIP in serum-starved glioma cells. J. Mol. Neurosci. 2013, 51, 503–513. [Google Scholar] [CrossRef]
- Xu, H.; Jia, J. Immune-Related Hub Genes and the Competitive Endogenous RNA Network in Alzheimer’s Disease. J. Alzheimers Dis. 2020, 77, 1255–1265. [Google Scholar] [CrossRef]
- Affymetrix Microarray Suite User Guide, 5th ed.; Affymetrix: Santa Clara, CA, USA, 2001.
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.H.; Chen, Y.C.; Yu, C.S.; Hwang, J.K. Predicting disulfide connectivity patterns. Proteins 2007, 67, 262–270. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.S.; Lu, C.H. Identification of antifreeze proteins and their functional residues by support vector machine and genetic algorithms based on n-peptide compositions. PLoS ONE 2011, 6, e20445. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Yu, C.S.; Wu, H.W.; Chang, Y.J.; Lin, C.P.; Lu, C.H. The structure-based cancer-related single amino acid variation prediction. Sci. Rep. 2021, 11, 13599. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, C.J. LIBSVM: A Library for Support Vector Machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Lin, C.-J. Formulations of support vector machines: A note from an optimization point of view. Neural Comput. 2001, 13, 307–317. [Google Scholar] [CrossRef]


| Datasets | Accuracy | Sensitivity | Specificity | MCC | Precision | F1 Score |
|---|---|---|---|---|---|---|
| GBM1 | 0.9176 | 0.9517 | 0.8648 | 0.8265 | 0.9156 | 0.9333 |
| GBM2 | 0.8835 | 0.8947 | 0.8722 | 0.7672 | 0.8755 | 0.8850 |
| Datasets | Overexpressed Genes | Underexpressed Genes |
|---|---|---|
| GBM1 | COL6A2, ABCC3, COL8A1, FAM20A, ADM1, CTHRC1 | FERMT1, LINC008362, DLL3, P2RY12, CHST9, IGFN11, CSDC2, ETNPPL, VIPR2, MGAT4C1, DLL1, TNR, GDF10 |
| GBM2 | PDPN, IBSP, MIR210HG2, ADM1, GPX8, MYL9, PDLIM4 | IRX2, SHANK2, IGFN11, MGAT4C1, ENHO, LUZP2, DPP10, CDHR1, AKR1C3, SCG3 |
| Gene | Encoded Protein | Reported Roles/Functions in Gliomas | References |
|---|---|---|---|
| ABCC3 | ATP-binding cassette subfamily C member 3 | Lower expression in GBM tissue versus normal brain tissue; low expression is associated with low survival rates. | Su et al., 2020 [38] |
| Upregulated expression correlates with poor overall survival in GBM. | Jiang et al., 2021 [10] | ||
| ADM | Proadrenomedullin | Positively regulated by STAT-3 signaling; enhances the migration of astroglioma cells. | Lim et al., 2014 [41] |
| COL6A2 | Collagen alpha-2(VI) chain | High expression associated with worse prognosis; induces tumor cell proliferation in recurrent and high-risk low-grade glioma. | Chen et al., 2020 [39] |
| COL8A1 | Collagen alpha-1(VIII) chain | High expression correlated with poor overall survival in GBM. | Jiang et al., 2021 [10] |
| CTHRC1 | Collagen triple helix repeat containing protein-1 | Increased expression in glioma tissue is associated with WHO disease stage; regulates tumor cell invasion, migration and adhesion. | Mei et al., 2017 [42] |
| FAM20A | Psedokinase FAM20A | Biomarker for low-grade glioma; overexpression predicts poor outcomes. | Feng et al., 2021 [40] |
| Associated with disrupted immune responses in the GBM microenvironment. | Du et al., 2020 [43] | ||
| GPX8 | Glutathione peroxidase 8 | N/A. | |
| IBSP | Integrin-binding bone sialoprotein 2 | High expression correlated with poor overall survival in GBM. | Jiang et al., 2021 [10] |
| MIR210HG | Nonprotein-coding gene | Identified as an EMT-related lncRNA in gliomas. | Tao et al., 2021 [44] |
| Serves as a biomarker for glioma diagnosis. | Min et al., 2016 [45] | ||
| MYL9 | Myosin regulatory light polypeptide 9 | High expression is associated with a poor prognosis and is increased in patients with recurrent disease. | Kruthika et al., 2019 [13] |
| The DAPK1-ITPRIP-MYL9 complex promotes the progression of malignant glioma. | Cao et al., 2021 [37] | ||
| PDLIM4 | PDZ and LIM domain protein 4 | Biomarker for high-grade glioma. | de Tayrac et al., 2013 [36] |
| PDPN | Podoplanin | Correlated with poor overall survival in GBM. | Jiang et al., 2021 [10] |
| Increases tumor cell migration and angiogenesis in malignant glioma. | Grau et al., 2015 [46] |
| Gene | Encoded Protein | Reported Roles/Functions in Gliomas | References |
|---|---|---|---|
| AKR1C3 | Aldo-keto reductase family 1 member C3 | A hormone activity regulator and prostaglandin F synthase that is expressed in GBM and oligodendrogliomas; associated with the duration of overall survival in patients with gliomas. | Park et al., 2010 [50] |
| CDHR1 | Cadherin-related family member 1 | Downregulated in GBM and other gliomas (compared with normal brain tissue); lower expression of CDHR1 is associated with worse clinical prognosis in GBM. | Wang et al., 2021 [24] |
| CHST9 | Carbohydrate sulfotransferase 9 | N/A. | |
| CSDC2 | Cold shock domain-containing protein C2 | N/A. | |
| DLL1 | Delta-like ligand 1 | Contributes to Notch signaling, which suppresses glioma stem cell differentiation and maintains their stem cell properties that contribute to GBM tumorigenesis. | Bazzoni et al., 2019 [20], Talukdar et al., 2016 [25] |
| DLL3 | Delta-like ligand 3 | An inhibitory ligand-driven activation of the Notch pathway and is a potent prognostic factor for malignant glioma; low DLL3 expression is linked to shorter overall survival. | Maimaiti et al., 2021 [26] |
| DPP10 | Inactive dipeptidyl peptidase 10 | Underexpressed in GBM but overexpressed in diffuse astrocytomas and anaplastic astrocytomas. | Gonzalez-Garcia et al., 2020 [51] |
| ENHO | Adropin (energy homeostasis-associated protein) | N/A. | |
| ETNPPL | Ethanolamine phosphate phospholyase | Underexpressed in GBM but overexpressed in diffuse astrocytomas and anaplastic astrocytomas. | Gonzalez-Garcia et al., 2020 [51] |
| FERMT1 | Fermitin family member 1 | N/A. | |
| GDF10 (BMP3b) | Growth differentiation factor 10 | Associated with progression-free survival in GBM in a gender-dependent manner (PFS probability falls faster in males with high GDF10 expression than in females). | Serao et al., 2011 [47] |
| IFGN1 | Immunoglobulin-like and fibronectin type III domain-containing protein 1 | N/A. | |
| IRX2 | Iroquois-class homeodomain protein IRX-2 | Biomarker of pilocytic astrocytoma localization. | Antonelli et al., 2018 [52] |
| LINC00836 | Long intergenic nonprotein-coding RNA 836 | N/A. | |
| LUZP2 | Leucine zipper protein 2 | Crucial for nervous system extracellular matrix development; downregulated expression corresponds with increasing tumor stage in low-grade gliomas. | Chen et al., 2020 [48] |
| MGAT4C | α-1,3-mannosyl-glycoprotein 4-β-N-acetylglucosaminyltransferase C | N/A. | |
| P2RY12 | P2Y purinoceptor 12 | A specific marker for resident microglia in gliomas; its expression and localization correspond with tumor stage and M1/M2 immune responses. | Zhu et al., 2017 [53] |
| SCG3 | Secretogranin III | Expression is inversely correlated with malignancy grade; high in oligodendrogliomas and low in GBM. | Wang et al., 2021 [27] |
| SHANK2 | SH3 and multiple ankyrin repeat domains 2 | N/A. | |
| TNR | Tenascin-R | Low expression in GBM; TNR dysregulation in GBM is associated with glioma malignancy. | Bi et al., 2017 [49] |
| VIPR2 | Vasoactive intestinal polypeptide receptor 2 | Overexpressed in gliomas, particularly in oligodendrogliomas. | Jaworski et al., 2000 [54] |
| Tumor Type | Number of Patients |
|---|---|
| Astrocytoma | 148 |
| Glioblastoma multiforme | 228 |
| Mixed | 11 |
| Oligodendroglioma | 67 |
| Unclassified 1 | 1 |
| Unknown 2 | 67 |
| Control 3 | 28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.-H.; Wei, S.-T.; Liu, J.-J.; Chang, Y.-J.; Lin, Y.-F.; Yu, C.-S.; Chang, S.L.-Y. Recognition of a Novel Gene Signature for Human Glioblastoma. Int. J. Mol. Sci. 2022, 23, 4157. https://doi.org/10.3390/ijms23084157
Lu C-H, Wei S-T, Liu J-J, Chang Y-J, Lin Y-F, Yu C-S, Chang SL-Y. Recognition of a Novel Gene Signature for Human Glioblastoma. International Journal of Molecular Sciences. 2022; 23(8):4157. https://doi.org/10.3390/ijms23084157
Chicago/Turabian StyleLu, Chih-Hao, Sung-Tai Wei, Jia-Jun Liu, Yu-Jen Chang, Yu-Feng Lin, Chin-Sheng Yu, and Sunny Li-Yun Chang. 2022. "Recognition of a Novel Gene Signature for Human Glioblastoma" International Journal of Molecular Sciences 23, no. 8: 4157. https://doi.org/10.3390/ijms23084157






