Salting-Out of DNA Origami Nanostructures by Ammonium Sulfate
Abstract
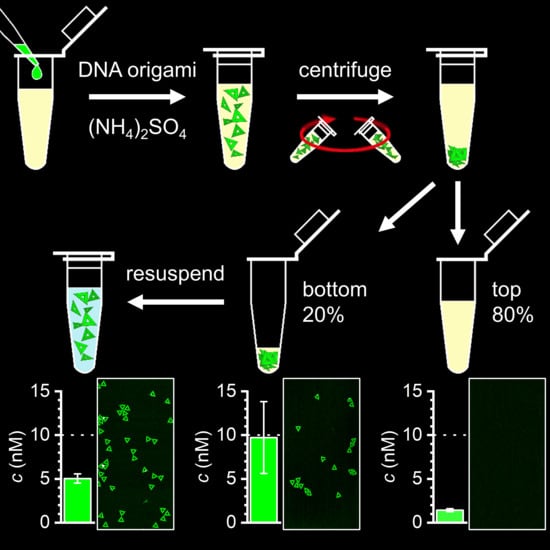
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. DNA Origami Synthesis and Purification
4.2. Salting-Out and Resuspension Experiments
4.3. Concentration Measurements
4.4. AFM Imaging
4.5. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramm, K.; Schröder, T.; Gouge, J.; Vera, A.M.; Gupta, K.; Heiss, F.B.; Liedl, T.; Engel, C.; Berger, I.; Vannini, A.; et al. DNA origami-based single-molecule force spectroscopy elucidates RNA Polymerase III pre-initiation complex stability. Nat. Commun. 2020, 11, 2828. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Le, J.V.; Crocker, K.; Darcy, M.A.; Halley, P.D.; Zhao, D.; Andrioff, N.; Croy, C.; Poirier, M.G.; Bundschuh, R.; et al. A nanoscale DNA force spectrometer capable of applying tension and compression on biomolecules. Nucleic Acids Res. 2021, 49, 8987–8999. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Tian, R.; Wu, J.; Liu, S.; Wang, Y.; Wen, M.; Shang, Y.; Liu, Q.; Li, Y.; Guo, Y.; et al. A DNA origami-based aptamer nanoarray for potent and reversible anticoagulation in hemodialysis. Nat. Commun. 2021, 12, 358. [Google Scholar] [CrossRef]
- Li, W.; Wang, C.; Lv, H.; Wang, Z.; Zhao, M.; Liu, S.; Gou, L.; Zhou, Y.; Li, J.; Zhang, J.; et al. A DNA Nanoraft-Based Cytokine Delivery Platform for Alleviation of Acute Kidney Injury. ACS Nano 2021, 15, 18237–18249. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, X.; Liu, P.; Wang, F.; Ariyama, H.; Ando, T.; Lin, J.; Wang, L.; Hu, J.; Li, B.; et al. Capturing transient antibody conformations with DNA origami epitopes. Nat. Commun. 2020, 11, 3114. [Google Scholar] [CrossRef]
- Lee, A.J.; Endo, M.; Hobbs, J.K.; Davies, A.G.; Wälti, C. Micro-homology intermediates: RecA’s transient sampling revealed at the single molecule level. Nucleic Acids Res. 2021, 49, 1426–1435. [Google Scholar] [CrossRef]
- Ketterer, P.; Ananth, A.N.; Laman Trip, D.S.; Mishra, A.; Bertosin, E.; Ganji, M.; van der Torre, J.; Onck, P.; Dietz, H.; Dekker, C. DNA origami scaffold for studying intrinsically disordered proteins of the nuclear pore complex. Nat. Commun. 2018, 9, 902. [Google Scholar] [CrossRef]
- Aksel, T.; Yu, Z.; Cheng, Y.; Douglas, S.M. Molecular goniometers for single-particle cryo-electron microscopy of DNA-binding proteins. Nat. Biotechnol. 2021, 39, 378–386. [Google Scholar] [CrossRef]
- Kielar, C.; Reddavide, F.V.; Tubbenhauer, S.; Cui, M.; Xu, X.; Grundmeier, G.; Zhang, Y.; Keller, A. Pharmacophore Nanoarrays on DNA Origami Substrates as a Single-Molecule Assay for Fragment-Based Drug Discovery. Angew. Chem. Int. Ed. Engl. 2018, 57, 14873–14877. [Google Scholar] [CrossRef]
- Huang, J.; Suma, A.; Cui, M.; Grundmeier, G.; Carnevale, V.; Zhang, Y.; Kielar, C.; Keller, A. Arranging Small Molecules with Subnanometer Precision on DNA Origami Substrates for the Single-Molecule Investigation of Protein–Ligand Interactions. Small Struct. 2020, 1, 2000038. [Google Scholar] [CrossRef]
- Pfeiffer, M.; Trofymchuk, K.; Ranallo, S.; Ricci, F.; Steiner, F.; Cole, F.; Glembockyte, V.; Tinnefeld, P. Single antibody detection in a DNA origami nanoantenna. iScience 2021, 24, 103072. [Google Scholar] [CrossRef] [PubMed]
- Tapio, K.; Mostafa, A.; Kanehira, Y.; Suma, A.; Dutta, A.; Bald, I. A Versatile DNA Origami-Based Plasmonic Nanoantenna for Label-Free Single-Molecule Surface-Enhanced Raman Spectroscopy. ACS Nano 2021, 15, 7065–7077. [Google Scholar] [CrossRef] [PubMed]
- Gwosch, K.C.; Pape, J.K.; Balzarotti, F.; Hoess, P.; Ellenberg, J.; Ries, J.; Hell, S.W. MINFLUX nanoscopy delivers 3D multicolor nanometer resolution in cells. Nat. Methods 2020, 17, 217–224. [Google Scholar] [CrossRef]
- Kielar, C.; Zhu, S.; Grundmeier, G.; Keller, A. Quantitative Assessment of Tip Effects in Single-Molecule High-Speed Atomic Force Microscopy Using DNA Origami Substrates. Angew. Chem. Int. Ed. Engl. 2020, 59, 14336–14341. [Google Scholar] [CrossRef]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.-Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Krainer, G.; Grundmeier, G.; Schlierf, M.; Keller, A. Structural stability of DNA origami nanostructures in the presence of chaotropic agents. Nanoscale 2016, 8, 10398–10405. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Da, Z.; Zhang, B.; Isbell, M.A.; Dong, Y.; Zhou, X.; Liu, H.; Heng, J.Y.Y.; Yang, Z. Stability study of tubular DNA origami in the presence of protein crystallisation buffer. RSC Adv. 2015, 5, 58734–58737. [Google Scholar] [CrossRef]
- Funke, J.J.; Dietz, H. Placing molecules with Bohr radius resolution using DNA origami. Nat. Nanotechnol. 2016, 11, 47–52. [Google Scholar] [CrossRef]
- Halvorsen, K.; Kizer, M.E.; Wang, X.; Chandrasekaran, A.R.; Basanta-Sanchez, M. Shear Dependent LC Purification of an Engineered DNA Nanoswitch and Implications for DNA Origami. Anal. Chem. 2017, 89, 5673–5677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; Perrault, S.D.; Kwak, M.; Graf, F.; Shih, W.M. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 2013, 41, e40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.; Benson, E.; Högberg, B. Purification of functionalized DNA origami nanostructures. ACS Nano 2015, 9, 4968–4975. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Teske, J.; Kemper, U.; Seidel, R. Sequential Pull-Down Purification of DNA Origami Superstructures. Small 2021, 17, 2007218. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.; Martin, T.G.; Praetorius, F.; Dietz, H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. Engl. 2014, 53, 12735–12740. [Google Scholar] [CrossRef] [Green Version]
- Bellot, G.; McClintock, M.A.; Lin, C.; Shih, W.M. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat. Methods 2011, 8, 192–194. [Google Scholar] [CrossRef]
- Ye, J.; Aftenieva, O.; Bayrak, T.; Jain, A.; König, T.A.F.; Erbe, A.; Seidel, R. Complex Metal Nanostructures with Programmable Shapes from Simple DNA Building Blocks. Adv. Mater. 2021, 33, 2100381. [Google Scholar] [CrossRef]
- Bayrak, T.; Helmi, S.; Ye, J.; Kauert, D.; Kelling, J.; Schönherr, T.; Weichelt, R.; Erbe, A.; Seidel, R. DNA-Mold Templated Assembly of Conductive Gold Nanowires. Nano Lett. 2018, 18, 2116–2123. [Google Scholar] [CrossRef]
- Engelen, W.; Sigl, C.; Kadletz, K.; Willner, E.M.; Dietz, H. Antigen-Triggered Logic-Gating of DNA Nanodevices. J. Am. Chem. Soc. 2021, 143, 21630–21636. [Google Scholar] [CrossRef]
- Sigl, C.; Willner, E.M.; Engelen, W.; Kretzmann, J.A.; Sachenbacher, K.; Liedl, A.; Kolbe, F.; Wilsch, F.; Aghvami, S.A.; Protzer, U.; et al. Programmable icosahedral shell system for virus trapping. Nat. Mater. 2021, 20, 1281–1289. [Google Scholar] [CrossRef]
- Zadegan, R.M.; Lindau, E.G.; Klein, W.P.; Green, C.; Graugnard, E.; Yurke, B.; Kuang, W.; Hughes, W.L. Twisting of DNA Origami from Intercalators. Sci. Rep. 2017, 7, 7382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liedl, T.; Högberg, B.; Tytell, J.; Ingber, D.E.; Shih, W.M. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 2010, 5, 520–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijäs, H.; Shen, B.; Heuer-Jungemann, A.; Keller, A.; Kostiainen, M.A.; Liedl, T.; Ihalainen, J.A.; Linko, V. Unraveling the interaction between doxorubicin and DNA origami nanostructures for customizable chemotherapeutic drug release. Nucleic Acids Res. 2021, 49, 3048–3062. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Chou, J.J.; Shih, W.M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl. Acad. Sci. USA 2007, 104, 6644–6648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Hartl, C.; Frank, K.; Heuer-Jungemann, A.; Fischer, S.; Nickels, P.C.; Nickel, B.; Liedl, T. 3D DNA Origami Crystals. Adv. Mater. 2018, 30, 1800273. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Kielar, C.; Zhu, S.; Sikeler, C.; Xu, X.; Möser, C.; Grundmeier, G.; Liedl, T.; Heuer-Jungemann, A.; Smith, D.M.; et al. Cryopreservation of DNA Origami Nanostructures. Small 2020, 16, 1905959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strauss, M.T.; Schueder, F.; Haas, D.; Nickels, P.C.; Jungmann, R. Quantifying absolute addressability in DNA origami with molecular resolution. Nat. Commun. 2018, 9, 1600. [Google Scholar] [CrossRef]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef]
- Ijäs, H.; Hakaste, I.; Shen, B.; Kostiainen, M.A.; Linko, V. Reconfigurable DNA Origami Nanocapsule for pH-Controlled Encapsulation and Display of Cargo. ACS Nano 2019, 13, 5959–5967. [Google Scholar] [CrossRef] [Green Version]
- Bastings, M.M.C.; Anastassacos, F.M.; Ponnuswamy, N.; Leifer, F.G.; Cuneo, G.; Lin, C.; Ingber, D.E.; Ryu, J.H.; Shih, W.M. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018, 18, 3557–3564. [Google Scholar] [CrossRef]
- Ingham, K.C. Precipitation of proteins with polyethylene glycol. In Guide to Protein Purification; Elsevier: Amsterdam, The Netherlands, 1990; pp. 301–306. ISBN 9780121820831. [Google Scholar]
- Hyde, A.M.; Zultanski, S.L.; Waldman, J.H.; Zhong, Y.-L.; Shevlin, M.; Peng, F. General Principles and Strategies for Salting-Out Informed by the Hofmeister Series. Org. Process. Res. Dev. 2017, 21, 1355–1370. [Google Scholar] [CrossRef] [Green Version]
- Lis, J.T.; Schleif, R. Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res. 1975, 2, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Pulleyblank, D.; Michalak, M.; Daisley, S.L.; Glick, R. A method for the purification of E. coli plasmid DNA by homogeneous lysis and polyethylene glycol precipitation. Mol. Biol. Rep. 1983, 9, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Atha, D.H.; Ingham, K.C. Mechanism of precipitation of proteins by polyethylene glycols. Analysis in terms of excluded volume. J. Biol. Chem. 1981, 256, 12108–12117. [Google Scholar] [CrossRef]
- Duong-Ly, K.C.; Gabelli, S.B. Salting out of proteins using ammonium sulfate precipitation. Methods Enzymol. 2014, 541, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, P. Protein Precipitation Using Ammonium Sulfate. Curr. Protoc. Protein Sci. 2016, 84, A.3F.1–A.3F.9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novák, P.; Havlíček, V. Protein Extraction and Precipitation. Proteomic Profiling and Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2016; pp. 51–62. ISBN 9780444636881. [Google Scholar]
- Freitas, S.S.; Santos, J.A.L.; Prazeres, D.M.F. Optimization of isopropanol and ammonium sulfate precipitation steps in the purification of plasmid DNA. Biotechnol. Prog. 2006, 22, 1179–1186. [Google Scholar] [CrossRef]
- Schneider, F.; Möritz, N.; Dietz, H. The sequence of events during folding of a DNA origami. Sci. Adv. 2019, 5, eaaw1412. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.E.; Dannenberg, F.; Ouldridge, T.E.; Kwiatkowska, M.; Turberfield, A.J.; Bath, J. Guiding the folding pathway of DNA origami. Nature 2015, 525, 82–86. [Google Scholar] [CrossRef]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem. Int. Ed. Engl. 2018, 57, 9470–9474. [Google Scholar] [CrossRef]
- Roodhuizen, J.A.L.; Hendrikx, P.J.T.M.; Hilbers, P.A.J.; de Greef, T.F.A.; Markvoort, A.J. Counterion-Dependent Mechanisms of DNA Origami Nanostructure Stabilization Revealed by Atomistic Molecular Simulation. ACS Nano 2019, 13, 10798–10809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Krainer, G.; Grundmeier, G.; Schlierf, M.; Keller, A. Cation-Induced Stabilization and Denaturation of DNA Origami Nanostructures in Urea and Guanidinium Chloride. Small 2017, 13, 1702100. [Google Scholar] [CrossRef]
- Bui, H.; Onodera, C.; Kidwell, C.; Tan, Y.; Graugnard, E.; Kuang, W.; Lee, J.; Knowlton, W.B.; Yurke, B.; Hughes, W.L. Programmable periodicity of quantum dot arrays with DNA origami nanotubes. Nano Lett. 2010, 10, 3367–3372. [Google Scholar] [CrossRef] [PubMed]
- Panja, A.S.; Maiti, S.; Bandyopadhyay, B. Protein stability governed by its structural plasticity is inferred by physicochemical factors and salt bridges. Sci. Rep. 2020, 10, 1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Singh, N. Effect of salt concentration on the stability of heterogeneous DNA. Physica A 2015, 419, 328–334. [Google Scholar] [CrossRef] [Green Version]
- Rathee, V.S.; Sidky, H.; Sikora, B.J.; Whitmer, J.K. Explicit Ion Effects on the Charge and Conformation of Weak Polyelectrolytes. Polymers 2019, 11, 183. [Google Scholar] [CrossRef] [Green Version]
- Rathee, V.S.; Sidky, H.; Sikora, B.J.; Whitmer, J.K. Role of Associative Charging in the Entropy-Energy Balance of Polyelectrolyte Complexes. J. Am. Chem. Soc. 2018, 140, 15319–15328. [Google Scholar] [CrossRef]
- Salis, A.; Ninham, B.W. Models and mechanisms of Hofmeister effects in electrolyte solutions, and colloid and protein systems revisited. Chem. Soc. Rev. 2014, 43, 7358–7377. [Google Scholar] [CrossRef] [Green Version]
- Asakura, S.; Oosawa, F. Interaction between particles suspended in solutions of macromolecules. J. Polym. Sci. 1958, 33, 183–192. [Google Scholar] [CrossRef]
- Duboué-Dijon, E.; Fogarty, A.C.; Hynes, J.T.; Laage, D. Dynamical Disorder in the DNA Hydration Shell. J. Am. Chem. Soc. 2016, 138, 7610–7620. [Google Scholar] [CrossRef]
- Opherden, L.; Oertel, J.; Barkleit, A.; Fahmy, K.; Keller, A. Paramagnetic decoration of DNA origami nanostructures by Eu3+ coordination. Langmuir 2014, 30, 8152–8159. [Google Scholar] [CrossRef] [PubMed]
- Kollmann, F.; Ramakrishnan, S.; Shen, B.; Grundmeier, G.; Kostiainen, M.A.; Linko, V.; Keller, A. Superstructure-Dependent Loading of DNA Origami Nanostructures with a Groove-Binding Drug. ACS Omega 2018, 3, 9441–9448. [Google Scholar] [CrossRef] [PubMed]
- Hanke, M.; Gonzalez Orive, A.; Grundmeier, G.; Keller, A. Effect of DNA Origami Nanostructures on hIAPP Aggregation. Nanomaterials 2020, 10, 2200. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Keller, A.; Linko, V. Real-Time Observation of Superstructure-Dependent DNA Origami Digestion by DNase I Using High-Speed Atomic Force Microscopy. ChemBioChem 2019, 20, 2818–2823. [Google Scholar] [CrossRef]
- Stopar, A.; Coral, L.; Di Giacomo, S.; Adedeji, A.F.; Castronovo, M. Binary control of enzymatic cleavage of DNA origami by structural antideterminants. Nucleic Acids Res. 2018, 46, 995–1006. [Google Scholar] [CrossRef] [Green Version]
- Suma, A.; Stopar, A.; Nicholson, A.W.; Castronovo, M.; Carnevale, V. Global and local mechanical properties control endonuclease reactivity of a DNA origami nanostructure. Nucleic Acids Res. 2020, 48, 4672–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for SPM data analysis. Open Physics 2012, 10, 181–188. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanke, M.; Hansen, N.; Chen, R.; Grundmeier, G.; Fahmy, K.; Keller, A. Salting-Out of DNA Origami Nanostructures by Ammonium Sulfate. Int. J. Mol. Sci. 2022, 23, 2817. https://doi.org/10.3390/ijms23052817
Hanke M, Hansen N, Chen R, Grundmeier G, Fahmy K, Keller A. Salting-Out of DNA Origami Nanostructures by Ammonium Sulfate. International Journal of Molecular Sciences. 2022; 23(5):2817. https://doi.org/10.3390/ijms23052817
Chicago/Turabian StyleHanke, Marcel, Niklas Hansen, Ruiping Chen, Guido Grundmeier, Karim Fahmy, and Adrian Keller. 2022. "Salting-Out of DNA Origami Nanostructures by Ammonium Sulfate" International Journal of Molecular Sciences 23, no. 5: 2817. https://doi.org/10.3390/ijms23052817