Granzymes: The Molecular Executors of Immune-Mediated Cytotoxicity
Abstract
:1. Introduction
2. Granzymes
3. Gzm Sequence and Phylogeny Analysis
4. Gzm Production and Regulation
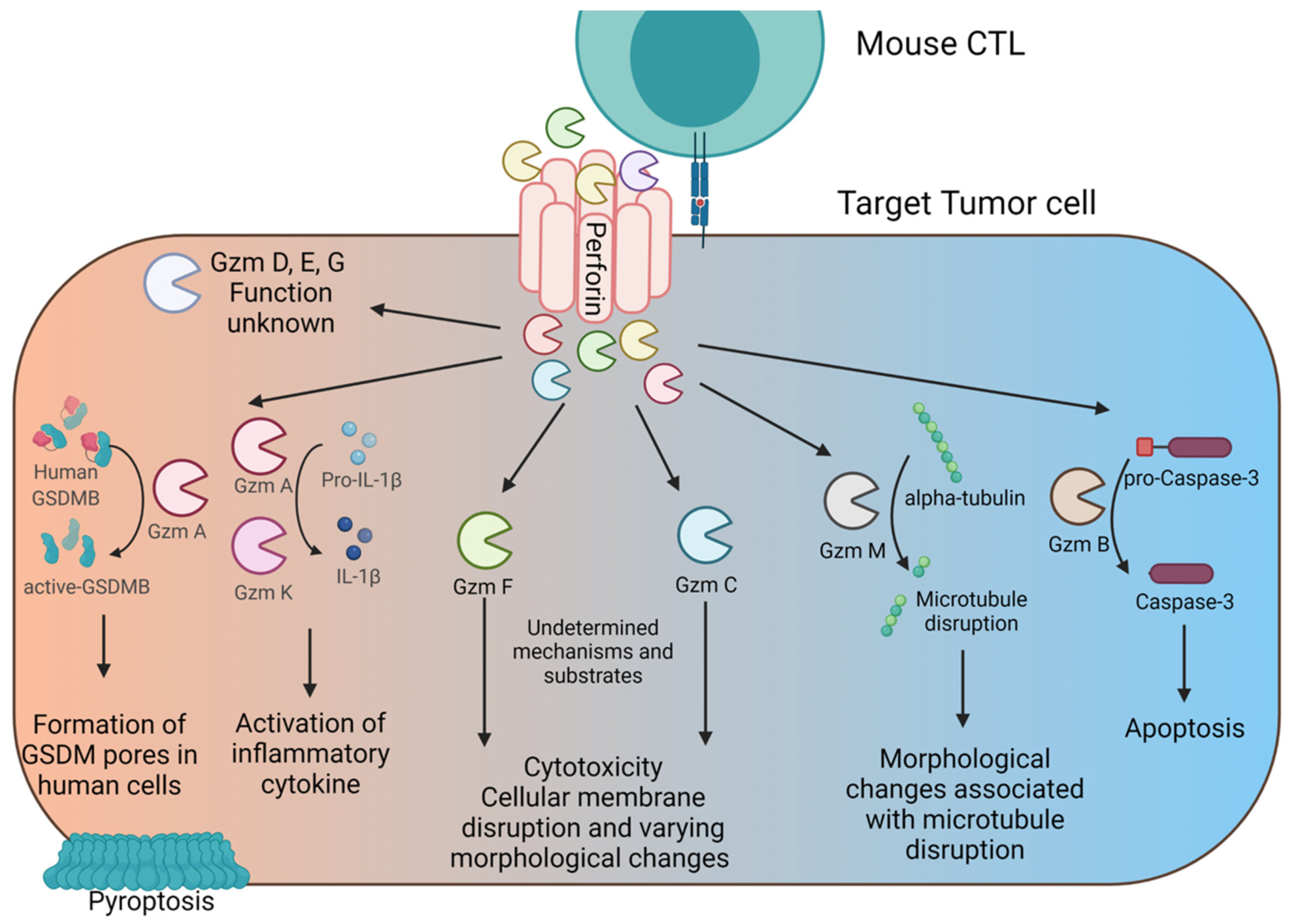
5. Cytotoxicity and Other Impacts of Gzms
5.1. Gzm B
5.2. Gzm A
5.3. Gzm M
5.4. Gzm K
5.5. Gzm F
5.6. Gzm C
5.7. Gzm H
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pardo, J.; Balkow, S.; Anel, A.; Simon, M.M. Granzymes are essential for natural killer cell-mediated and perf-facilitated tumor control. Eur. J. Immunol. 2002, 32, 2881–2887. [Google Scholar] [CrossRef]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef]
- Pardo, J.; Wallich, R.; Ebnet, K.; Iden, S.; Zentgraf, H.; Martin, P.; Ekiciler, A.; Prins, A.; Müllbacher, A.; Huber, M.; et al. Granzyme B is expressed in mouse mast cells in vivo and in vitro and causes delayed cell death independent of perforin. Cell Death Differ. 2007, 14, 1768–1779. [Google Scholar] [CrossRef] [PubMed]
- Buchan, S.L.; Fallatah, M.; Thirdborough, S.M.; Taraban, V.Y.; Rogel, A.; Thomas, L.J.; Penfold, C.A.; He, L.Z.; Curran, M.A.; Keler, T.; et al. PD-1 Blockade and CD27 Stimulation Activate Distinct Transcriptional Programs That Synergize for CD8(+) T-Cell-Driven Antitumor Immunity. Clin. Cancer Res. 2018, 24, 2383–2394. [Google Scholar] [CrossRef] [Green Version]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Hur, J.; Yang, H.M.; Yoon, C.H.; Lee, C.S.; Park, K.W.; Kim, J.H.; Kim, T.Y.; Kim, J.Y.; Kang, H.J.; Chae, I.H.; et al. Identification of a novel role of T cells in postnatal vasculogenesis: Characterization of endothelial progenitor cell colonies. Circulation 2007, 116, 1671–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raphael, I.; Nalawade, S.; Eagar, T.N.; Forsthuber, T.G. T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 2015, 74, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Zlotnik, A.; Yoshie, O. Chemokines: A New Classification System and Their Role in Immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef] [Green Version]
- Waring, P.; Müllbacher, A. Cell death induced by the Fas/Fas ligand pathway and its role in pathology. Immunol. Cell Biol. 1999, 77, 312–317. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Dixit, V.M. Death receptors: Signaling and modulation. Science 1998, 281, 1305–1308. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazi, A.; Dixit, V.M. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 1999, 11, 255–260. [Google Scholar] [CrossRef]
- Bullani, R.R.; Wehrli, P.; Viard-Leveugle, I.; Rimoldi, D.; Cerottini, J.C.; Saurat, J.H.; Tschopp, J.; French, L.E. Frequent downregulation of Fas (CD95) expression and function in melanoma. Melanoma Res. 2002, 12, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Horvath, C.M.; Wen, Z.; Schreiber, R.D.; Darnell, J.E. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. USA 1996, 93, 7673–7678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Fu, X.Y.; Plate, J.; Chong, A.S. IFN-gamma induces cell growth inhibition by Fas-mediated apoptosis: Requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 1998, 58, 2832–2837. [Google Scholar] [PubMed]
- Takeda, K.; Smyth, M.J.; Cretney, E.; Hayakawa, Y.; Kayagaki, N.; Yagita, H.; Okumura, K. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J. Exp. Med. 2002, 195, 161–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulda, S.; Debatin, K.-M. IFNγ sensitizes for apoptosis by upregulating caspase-8 expression through the Stat1 pathway. Oncogene 2002, 21, 2295–2308. [Google Scholar] [CrossRef] [Green Version]
- Chin, Y.E.; Kitagawa, M.; Kuida, K.; Flavell, R.A.; Fu, X.Y. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 1997, 17, 5328–5337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kammertoens, T.; Friese, C.; Arina, A.; Idel, C.; Briesemeister, D.; Rothe, M.; Ivanov, A.; Szymborska, A.; Patone, G.; Kunz, S.; et al. Tumour ischaemia by interferon-γ resembles physiological blood vessel regression. Nature 2017, 545, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Lostao, L.; Anel, A.; Pardo, J. How Do Cytotoxic Lymphocytes Kill Cancer Cells? Clin. Cancer Res. 2015, 21, 5047–5056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bots, M.; Medema, J.P. Granzymes at a glance. J. Cell Sci. 2006, 119, 5011–5014. [Google Scholar] [CrossRef] [Green Version]
- Jenne, D.E.; Tschopp, J. Granzymes, a family of serine proteases released from granules of cytolytic T lymphocytes upon T cell receptor stimulation. Immunol. Rev. 1988, 103, 53–71. [Google Scholar] [CrossRef]
- Trapani, J.A.; Browne, K.A.; Dawson, M.; Smyth, M.J. Immunopurification of functional Asp-ase (natural killer cell granzyme B) using a monoclonal antibody. Biochem. Biophys. Res. Commun. 1993, 195, 910–920. [Google Scholar] [CrossRef]
- Dotiwala, F.; Mulik, S.; Polidoro, R.B.; Ansara, J.A.; Burleigh, B.A.; Walch, M.; Gazzinelli, R.T.; Lieberman, J. Killer lymphocytes use granulysin, perforin and granzymes to kill intracellular parasites. Nat. Med. 2016, 22, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Ernst, W.A.; Thoma-Uszynski, S.; Teitelbaum, R.; Ko, C.; Hanson, D.A.; Clayberger, C.; Krensky, A.M.; Leippe, M.; Bloom, B.R.; Ganz, T.; et al. Granulysin, a T Cell Product, Kills Bacteria by Altering Membrane Permeability. J. Immunol. 2000, 165, 7102–7108. [Google Scholar] [CrossRef] [Green Version]
- Stenger, S.; Hanson Dennis, A.; Teitelbaum, R.; Dewan, P.; Niazi Kayvan, R.; Froelich Christopher, J.; Ganz, T.; Thoma-Uszynski, S.; Melián, A.n.; Bogdan, C.; et al. An Antimicrobial Activity of Cytolytic T Cells Mediated by Granulysin. Science 1998, 282, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Law, R.H.; Lukoyanova, N.; Voskoboinik, I.; Caradoc-Davies, T.T.; Baran, K.; Dunstone, M.A.; D’Angelo, M.E.; Orlova, E.V.; Coulibaly, F.; Verschoor, S.; et al. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 2010, 468, 447–451. [Google Scholar] [CrossRef]
- Lichtenheld, M.G.; Olsen, K.J.; Lu, P.; Lowrey, D.M.; Hameed, A.; Hengartner, H.; Podack, E.R. Structure and function of human perforin. Nature 1988, 335, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Osińska, I.; Popko, K.; Demkow, U. Perforin: An important player in immune response. Cent Eur. J. Immunol. 2014, 39, 109–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odake, S.; Kam, C.M.; Narasimhan, L.; Poe, M.; Blake, J.T.; Krahenbuhl, O.; Tschopp, J.; Powers, J.C. Human and murine cytotoxic T lymphocyte serine proteases: Subsite mapping with peptide thioester substrates and inhibition of enzyme activity and cytolysis by isocoumarins. Biochemistry 1991, 30, 2217–2227. [Google Scholar] [CrossRef]
- Chowdhury, D.; Lieberman, J. Death by a thousand cuts: Granzyme pathways of programmed cell death. Annu. Rev. Immunol. 2008, 26, 389–420. [Google Scholar] [CrossRef] [Green Version]
- Poe, M.; Blake, J.T.; Boulton, D.A.; Gammon, M.; Sigal, N.H.; Wu, J.K.; Zweerink, H.J. Human cytotoxic lymphocyte granzyme B. Its purification from granules and the characterization of substrate and inhibitor specificity. J. Biol. Chem. 1991, 266, 98–103. [Google Scholar] [CrossRef]
- Turner, C.T.; Zeglinski, M.R.; Richardson, K.C.; Santacruz, S.; Hiroyasu, S.; Wang, C.; Zhao, H.; Shen, Y.; Sehmi, R.; Lima, H.; et al. Granzyme B Contributes to Barrier Dysfunction in Oxazolone-Induced Skin Inflammation through E-Cadherin and FLG Cleavage. J. Investig. Dermatol. 2021, 141, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Hiroyasu, S.; Hiroyasu, A.; Granville, D.J.; Tsuruta, D. Pathological functions of granzyme B in inflammatory skin diseases. J. Dermatol. Sci. 2021, 104, 76–82. [Google Scholar] [CrossRef]
- Zeglinski, M.R.; Granville, D.J. Granzymes in cardiovascular injury and disease. Cell. Signal. 2020, 76, 109804. [Google Scholar] [CrossRef] [PubMed]
- Thomas, H.E.; Trapani, J.A.; Kay, T.W.H. The role of perforin and granzymes in diabetes. Cell Death Differ. 2010, 17, 577–585. [Google Scholar] [CrossRef] [Green Version]
- Garzón-Tituaña, M.; Arias, M.A.; Sierra-Monzón, J.L.; Morte-Romea, E.; Santiago, L.; Ramirez-Labrada, A.; Martinez-Lostao, L.; Paño-Pardo, J.R.; Galvez, E.M.; Pardo, J. The Multifaceted Function of Granzymes in Sepsis: Some Facts and a Lot to Discover. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Hiroyasu, S.; Zeglinski, M.R.; Zhao, H.; Pawluk, M.A.; Turner, C.T.; Kasprick, A.; Tateishi, C.; Nishie, W.; Burleigh, A.; Lennox, P.A.; et al. Granzyme B inhibition reduces disease severity in autoimmune blistering diseases. Nat. Commun. 2021, 12, 302. [Google Scholar] [CrossRef]
- Baker, E.; Sayers, T.J.; Sutherland, G.R.; Smyth, M.J. The genes encoding NK cell granule serine proteases, human tryptase-2 (TRYP2) and human granzyme A (HFSP), both map to chromosome 5q11-q12 and define a new locus for cytotoxic lymphocyte granule tryptases. Immunogenetics 1994, 40, 235–237. [Google Scholar] [CrossRef]
- Yang, J.; Vrettou, C.; Connelley, T.; Morrison, W.I. Identification and annotation of bovine granzyme genes reveals a novel granzyme encoded within the trypsin-like locus. Immunogenetics 2018, 70, 585–597. [Google Scholar] [CrossRef] [Green Version]
- Trapani, J.A. Granzymes: A family of lymphocyte granule serine proteases. Genome Biol. 2001, 2, Reviews3014. [Google Scholar] [CrossRef]
- Li, X. The inducers of immunogenic cell death for tumor immunotherapy. Tumori 2018, 104, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Serrano-del Valle, A.; Anel, A.; Naval, J.; Marzo, I. Immunogenic Cell Death and Immunotherapy of Multiple Myeloma. Front. Cell Dev. Biol. 2019, 7. [Google Scholar] [CrossRef]
- Galluzzi, L.; Garg, A.D. Immunology of Cell Death in Cancer Immunotherapy. Cells 2021, 10, 1208. [Google Scholar] [CrossRef]
- Messmer, M.N.; Snyder, A.G.; Oberst, A. Comparing the effects of different cell death programs in tumor progression and immunotherapy. Cell Death Differ. 2019, 26, 115–129. [Google Scholar] [CrossRef]
- Van Hoecke, L.; Van Lint, S.; Roose, K.; Van Parys, A.; Vandenabeele, P.; Grooten, J.; Tavernier, J.; De Koker, S.; Saelens, X. Treatment with mRNA coding for the necroptosis mediator MLKL induces antitumor immunity directed against neo-epitopes. Nat. Commun. 2018, 9, 3417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, S.P.; Adrain, C.; Lüthi, A.U.; Duriez, P.J.; Martin, S.J. Human and murine granzyme B exhibit divergent substrate preferences. J. Cell Biol. 2007, 176, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; He, H.; Wang, K.; Shi, X.; Wang, Y.; Su, Y.; Wang, Y.; Li, D.; Liu, W.; Zhang, Y.; et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 2020, 368, eaaz7548. [Google Scholar] [CrossRef]
- Beresford, P.J.; Kam, C.M.; Powers, J.C.; Lieberman, J. Recombinant human granzyme A binds to two putative HLA-associated proteins and cleaves one of them. Proc. Natl. Acad. Sci. USA 1997, 94, 9285–9290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.; Beresford, P.J.; Oh, D.Y.; Zhang, D.; Lieberman, J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 2003, 112, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Irmler, M.; Hertig, S.; MacDonald, H.R.; Sadoul, R.; Becherer, J.D.; Proudfoot, A.; Solari, R.; Tschopp, J. Granzyme A is an interleukin 1 beta-converting enzyme. J. Exp. Med. 1995, 181, 1917–1922. [Google Scholar] [CrossRef] [Green Version]
- Martinvalet, D.; Dykxhoorn, D.M.; Ferrini, R.; Lieberman, J. Granzyme A Cleaves a Mitochondrial Complex I Protein to Initiate Caspase-Independent Cell Death. Cell 2008, 133, 681–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.; Beresford, P.J.; Zhang, D.; Lieberman, J. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell. Biol. 2002, 22, 2810–2820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.; Beresford, P.J.; Zhang, D.; Xu, Z.; Novina, C.D.; Yoshida, A.; Pommier, Y.; Lieberman, J. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat. Immunol. 2003, 4, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.; Scorrano, L.; Korsmeyer, S.J.; Ley, T.J. Cell death induced by granzyme C. Blood 2003, 101, 3093–3101. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Wu, L.; Wang, S.; Fan, Z. Granzyme F induces a novel death pathway characterized by Bid-independent cytochrome c release without caspase activation. Cell Death Differ. 2009, 16, 1694–1706. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Yu, J.; Zhang, L. Necroptosis: An alternative cell death program defending against cancer. Biochim. Biophys. Acta 2016, 1865, 228–236. [Google Scholar] [CrossRef] [Green Version]
- Leist, M.; Single, B.; Castoldi, A.F.; Kühnle, S.; Nicotera, P. Intracellular adenosine triphosphate (ATP) concentration: A switch in the decision between apoptosis and necrosis. J. Exp. Med. 1997, 185, 1481–1486. [Google Scholar] [CrossRef]
- Bovenschen, N.; Quadir, R.; van den Berg, A.L.; Brenkman, A.B.; Vandenberghe, I.; Devreese, B.; Joore, J.; Kummer, J.A. Granzyme K Displays Highly Restricted Substrate Specificity That Only Partially Overlaps with Granzyme A*. J. Biol. Chem. 2009, 284, 3504–3512. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.; Zhang, H.; Guo, Y.; Zhang, Q.; Hua, G.; Lu, H.; Hou, Q.; Liu, H.; Fan, Z. Granzyme K cleaves the nucleosome assembly protein SET to induce single-stranded DNA nicks of target cells. Cell Death Differ. 2007, 14, 489–499. [Google Scholar] [CrossRef]
- Plasman, K.; Demol, H.; Bird, P.I.; Gevaert, K.; Van Damme, P. Substrate Specificities of the Granzyme Tryptases A and K. J. Proteome Res. 2014, 13, 6067–6077. [Google Scholar] [CrossRef]
- Bouwman, A.C.; van Daalen, K.R.; Crnko, S.; ten Broeke, T.; Bovenschen, N. Intracellular and Extracellular Roles of Granzyme K. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chen, J.; Zhao, T.; Fan, Z. Granzyme K degrades the redox/DNA repair enzyme Ape1 to trigger oxidative stress of target cells leading to cytotoxicity. Mol. Immunol. 2008, 45, 2225–2235. [Google Scholar] [CrossRef] [PubMed]
- Cullen, S.P.; Afonina, I.S.; Donadini, R.; Lüthi, A.U.; Medema, J.P.; Bird, P.I.; Martin, S.J. Nucleophosmin is cleaved and inactivated by the cytotoxic granule protease granzyme M during natural killer cell-mediated killing. J. Biol. Chem. 2009, 284, 5137–5147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Xia, P.; Shi, L.; Fan, Z. FADD cleavage by NK cell granzyme M enhances its self-association to facilitate procaspase-8 recruitment for auto-processing leading to caspase cascade. Cell Death Differ. 2012, 19, 605–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, D.; Liu, S.; Shi, L.; Li, C.; Wu, L.; Fan, Z. Cleavage of survivin by Granzyme M triggers degradation of the survivin-X-linked inhibitor of apoptosis protein (XIAP) complex to free caspase activity leading to cytolysis of target tumor cells. J. Biol. Chem. 2010, 285, 18326–18335. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Hou, Q.; Zhao, T.; Zhang, H.; Zhang, Q.; Wu, L.; Fan, Z. Granzyme M directly cleaves inhibitor of caspase-activated DNase (CAD) to unleash CAD leading to DNA fragmentation. J. Immunol. 2006, 177, 1171–1178. [Google Scholar] [CrossRef] [Green Version]
- de Poot, S.A.H.; Westgeest, M.; Hostetter, D.R.; van Damme, P.; Plasman, K.; Demeyer, K.; Broekhuizen, R.; Gevaert, K.; Craik, C.S.; Bovenschen, N. Human and mouse granzyme M display divergent and species-specific substrate specificities. Biochem. J. 2011, 437, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Takano, N.; Matusi, H.; Takahashi, T. Granzyme N, a Novel Granzyme, Is Expressed in Spermatocytes and Spermatids of the Mouse Testis1. Biol. Reprod. 2004, 71, 1785–1795. [Google Scholar] [CrossRef]
- Caputo, A.; Garner, R.S.; Winkler, U.; Hudig, D.; Bleackley, R.C. Activation of recombinant murine cytotoxic cell proteinase-1 requires deletion of an amino-terminal dipeptide. J. Biol. Chem. 1993, 268, 17672–17675. [Google Scholar] [CrossRef]
- Dikov, M.M.; Springman, E.B.; Yeola, S.; Serafin, W.E. Processing of procarboxypeptidase A and other zymogens in murine mast cells. J. Biol. Chem. 1994, 269, 25897–25904. [Google Scholar] [CrossRef]
- Smyth, M.J.; McGuire, M.J.; Thia, K.Y. Expression of recombinant human granzyme B. A processing and activation role for dipeptidyl peptidase I. J. Immunol. 1995, 154, 6299–6305. [Google Scholar]
- Hedstrom, L. Serine protease mechanism and specificity. Chemical Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef]
- Sattar, R.; Ali, S.A.; Abbasi, A. Bioinformatics of granzymes: Sequence comparison and structural studies on granzyme family by homology modeling. Biochem. Biophys. Res. Commun. 2003, 308, 726–735. [Google Scholar] [CrossRef]
- Sutton, V.R.; Waterhouse, N.J.; Browne, K.A.; Sedelies, K.; Ciccone, A.; Anthony, D.; Koskinen, A.; Mullbacher, A.; Trapani, J.A. Residual active granzyme B in cathepsin C-null lymphocytes is sufficient for perforin-dependent target cell apoptosis. J. Cell Biol. 2007, 176, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Grossman, W.J.; Revell, P.A.; Lu, Z.H.; Johnson, H.; Bredemeyer, A.J.; Ley, T.J. The orphan granzymes of humans and mice. Curr. Opin. Immunol. 2003, 15, 544–552. [Google Scholar] [CrossRef]
- Sedelies, K.A.; Sayers, T.J.; Edwards, K.M.; Chen, W.; Pellicci, D.G.; Godfrey, D.I.; Trapani, J.A. Discordant regulation of granzyme H and granzyme B expression in human lymphocytes. J. Biol. Chem. 2004, 279, 26581–26587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, S.F.; Fehniger, T.A.; Dai, X.; Cao, X.; Ley, T.J. Differential Regulation of Granzyme B and C Expression in Murine Cytotoxic T and NK Cells. Blood 2007, 110, 2291. [Google Scholar] [CrossRef]
- Waugh, K.A.; Leach, S.M.; Moore, B.L.; Bruno, T.C.; Buhrman, J.D.; Slansky, J.E. Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model. J. Immunol. 2016, 197, 1477–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arina, A.; Beckett, M.; Fernandez, C.; Zheng, W.; Pitroda, S.; Chmura, S.J.; Luke, J.J.; Forde, M.; Hou, Y.; Burnette, B.; et al. Tumor-reprogrammed resident T cells resist radiation to control tumors. Nat. Commun. 2019, 10, 3959. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Sanz, J.A.; MacDonald, H.R.; Jenne, D.E.; Tschopp, J.; Nabholz, M. Cell specificity of granzyme gene expression. J. Immunol. 1990, 145, 3111–3118. [Google Scholar]
- Lieberman, J. The ABCs of granule-mediated cytotoxicity: New weapons in the arsenal. Nat. Rev. Immunol. 2003, 3, 361–370. [Google Scholar] [CrossRef]
- Matsubara, J.A.; Tian, Y.; Cui, J.Z.; Zeglinski, M.R.; Hiroyasu, S.; Turner, C.T.; Granville, D.J. Retinal Distribution and Extracellular Activity of Granzyme B: A Serine Protease That Degrades Retinal Pigment Epithelial Tight Junctions and Extracellular Matrix Proteins. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Buzza, M.S.; Zamurs, L.; Sun, J.; Bird, C.H.; Smith, A.I.; Trapani, J.A.; Froelich, C.J.; Nice, E.C.; Bird, P.I. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J. Biol. Chem. 2005, 280, 23549–23558. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Merkulova, Y.; Raithatha, S.; Parkinson, L.G.; Shen, Y.; Cooper, D.; Granville, D.J. Extracellular granzyme K mediates endothelial activation through the cleavage of protease-activated receptor-1. FEBS J. 2016, 283, 1734–1747. [Google Scholar] [CrossRef] [Green Version]
- Heusel, J.W.; Wesselschmidt, R.L.; Shresta, S.; Russell, J.H.; Ley, T.J. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell 1994, 76, 977–987. [Google Scholar] [CrossRef]
- Hagn, M.; Sutton, V.R.; Trapani, J.A. A colorimetric assay that specifically measures Granzyme B proteolytic activity: Hydrolysis of Boc-Ala-Ala-Asp-S-Bzl. J. Vis. Exp. 2014, 93, e52419. [Google Scholar] [CrossRef] [Green Version]
- Pham, C.T.; Thomas, D.A.; Mercer, J.D.; Ley, T.J. Production of fully active recombinant murine granzyme B in yeast. J. Biol. Chem. 1998, 273, 1629–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Bird, C.H.; Buzza, M.S.; McKee, K.E.; Whisstock, J.C.; Bird, P.I. Expression and purification of recombinant human granzyme B from Pichia pastoris. Biochem. Biophys. Res. Commun. 1999, 261, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Xu, X.; Ewen, C.; Bleackley, R.C.; Kane, K.P. Isolation and characterization of novel single-chain Fv specific for human granzyme B. Hybrid. Hybridomics 2004, 23, 219–231. [Google Scholar] [CrossRef]
- Andrade, F.; Roy, S.; Nicholson, D.; Thornberry, N.; Rosen, A.; Casciola-Rosen, L. Granzyme B directly and efficiently cleaves several downstream caspase substrates: Implications for CTL-induced apoptosis. Immunity 1998, 8, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Stennicke, H.R.; Wang, B.; Green, D.R.; Janicke, R.U.; Srinivasan, A.; Seth, P.; Salvesen, G.S.; Froelich, C.J. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J. Biol. Chem. 1998, 273, 34278–34283. [Google Scholar] [CrossRef] [Green Version]
- Sutton, V.R.; Davis, J.E.; Cancilla, M.; Johnstone, R.W.; Ruefli, A.A.; Sedelies, K.; Browne, K.A.; Trapani, J.A. Initiation of apoptosis by granzyme B requires direct cleavage of bid, but not direct granzyme B-mediated caspase activation. J. Exp. Med. 2000, 192, 1403–1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julien, O.; Wells, J.A. Caspases and their substrates. Cell Death Dis. 2017, 24, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, D.; Takahama, S.; Shimizu, K.; Hayashi, S.; Endo, Y.; Sawasaki, T. Characterization of a caspase-3-substrate kinome using an N- and C-terminally tagged protein kinase library produced by a cell-free system. Cell Death Dis. 2010, 1, e89. [Google Scholar] [CrossRef]
- Wickman, G.R.; Julian, L.; Mardilovich, K.; Schumacher, S.; Munro, J.; Rath, N.; Zander, S.A.; Mleczak, A.; Sumpton, D.; Morrice, N.; et al. Blebs produced by actin–myosin contraction during apoptosis release damage-associated molecular pattern proteins before secondary necrosis occurs. Cell Death Dis. 2013, 20, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, T.A.; Herndon, J.; Elzey, B.; Griffith, T.S.; Schoenberger, S.; Green, D.R. Uptake of apoptotic antigen-coupled cells by lymphoid dendritic cells and cross-priming of CD8(+) T cells produce active immune unresponsiveness. J. Immunol. 2002, 168, 5589–5595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadok, V.A.; Bratton, D.L.; Konowal, A.; Freed, P.W.; Westcott, J.Y.; Henson, P.M. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Investig. 1998, 101, 890–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Li, F.; Liu, X.; Li, W.; Shi, W.; Liu, F.F.; O’Sullivan, B.; He, Z.; Peng, Y.; Tan, A.C.; et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 2011, 17, 860–866. [Google Scholar] [CrossRef]
- Bengsch, B.; Ohtani, T.; Herati, R.S.; Bovenschen, N.; Chang, K.M.; Wherry, E.J. Deep immune profiling by mass cytometry links human T and NK cell differentiation and cytotoxic molecule expression patterns. J. Immunol. Methods 2018, 453, 3–10. [Google Scholar] [CrossRef]
- Zou, H.; Li, Y.; Liu, X.; Wang, X. An APAF-1·Cytochrome c Multimeric Complex Is a Functional Apoptosome That Activates Procaspase-9*. J. Biol. Chem. 1999, 274, 11549–11556. [Google Scholar] [CrossRef] [Green Version]
- Saleh, A.; Srinivasula, S.M.; Acharya, S.; Fishel, R.; Alnemri, E.S. Cytochrome c and dATP-mediated Oligomerization of Apaf-1 Is a Prerequisite for Procaspase-9 Activation*. J. Biol. Chem. 1999, 274, 17941–17945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Benedict, M.A.; Ding, L.; Núñez, G. Role of cytochrome c and dATP/ATP hydrolysis in Apaf-1-mediated caspase-9 activation and apoptosis. EMBO J. 1999, 18, 3586–3595. [Google Scholar] [CrossRef] [PubMed]
- Cain, K.; Brown, D.G.; Langlais, C.; Cohen, G.M. Caspase Activation Involves the Formation of the Aposome, a Large (∼700 kDa) Caspase-activating Complex*. J. Biol. Chem. 1999, 274, 22686–22692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, J.A.; Prow, N.A.; Schroder, W.A.; Ellis, J.J.; Cumming, H.E.; Gearing, L.J.; Poo, Y.S.; Taylor, A.; Hertzog, P.J.; Di Giallonardo, F.; et al. RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathogens 2017, 13, e1006155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metkar, S.S.; Menaa, C.; Pardo, J.; Wang, B.; Wallich, R.; Freudenberg, M.; Kim, S.; Raja, S.M.; Shi, L.; Simon, M.M.; et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity 2008, 29, 720–733. [Google Scholar] [CrossRef] [Green Version]
- Aglietti, R.A.; Estevez, A.; Gupta, A.; Ramirez, M.G.; Liu, P.S.; Kayagaki, N.; Ciferri, C.; Dixit, V.M.; Dueber, E.C. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 2016, 113, 7858–7863. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Duncan, J.A.; Canna, S.W. The NLRC4 Inflammasome. Immunol. Rev. 2018, 281, 115–123. [Google Scholar] [CrossRef]
- Sollberger, G.; Strittmatter, G.E.; Garstkiewicz, M.; Sand, J.; Beer, H.D. Caspase-1: The inflammasome and beyond. Innate Immun. 2014, 20, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Martinvalet, D.; Zhu, P.; Lieberman, J. Granzyme A induces caspase-independent mitochondrial damage, a required first step for apoptosis. Immunity 2005, 22, 355–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, P.; Zhang, D.; Chowdhury, D.; Martinvalet, D.; Keefe, D.; Shi, L.; Lieberman, J. Granzyme A, which causes single-stranded DNA damage, targets the double-strand break repair protein Ku70. EMBO Rep. 2006, 7, 431–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, J.M.; Waterhouse, N.J.; Cretney, E.; Browne, K.A.; Ellis, S.; Trapani, J.A.; Smyth, M.J. Granzyme M mediates a novel form of perforin-dependent cell death. J. Biol. Chem. 2004, 279, 22236–22242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muppidi, J.R.; Lobito, A.A.; Ramaswamy, M.; Yang, J.K.; Wang, L.; Wu, H.; Siegel, R.M. Homotypic FADD interactions through a conserved RXDLL motif are required for death receptor-induced apoptosis. Cell Death Differ. 2006, 13, 1641–1650. [Google Scholar] [CrossRef]
- Pegram, H.J.; Haynes, N.M.; Smyth, M.J.; Kershaw, M.H.; Darcy, P.K. Characterizing the anti-tumor function of adoptively transferred NK cells in vivo. Cancer Immunol. Immunother. 2010, 59, 1235–1246. [Google Scholar] [CrossRef]
- Bovenschen, N.; de Koning, P.J.A.; Quadir, R.; Broekhuizen, R.; Damen, J.M.A.; Froelich, C.J.; Slijper, M.; Kummer, J.A. NK Cell Protease Granzyme M Targets α-Tubulin and Disorganizes the Microtubule Network. J. Immunol. 2008, 180, 8184–8191. [Google Scholar] [CrossRef]
- Zhao, T.; Zhang, H.; Guo, Y.; Fan, Z. Granzyme K directly processes bid to release cytochrome c and endonuclease G leading to mitochondria-dependent cell death. J. Biol. Chem. 2007, 282, 12104–12111. [Google Scholar] [CrossRef] [Green Version]
- Joeckel, L.T.; Wallich, R.; Martin, P.; Sanchez-Martinez, D.; Weber, F.C.; Martin, S.F.; Borner, C.; Pardo, J.; Froelich, C.; Simon, M.M. Mouse granzyme K has pro-inflammatory potential. Cell Death Differ. 2011, 18, 1112–1119. [Google Scholar] [CrossRef]
- Green, D.R.; Ferguson, T.; Zitvogel, L.; Kroemer, G. Immunogenic and tolerogenic cell death. Nat. Rev. Immunol. 2009, 9, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, A.; Vandenabeele, P.; Krysko, D.V. Necroptosis: The release of damage-associated molecular patterns and its physiological relevance. Immunity 2013, 38, 209–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fellows, E.; Gil-Parrado, S.; Jenne, D.E.; Kurschus, F.C. Natural killer cell–derived human granzyme H induces an alternative, caspase-independent cell-death program. Blood 2007, 110, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Zhao, T.; Zhang, H.; Lu, H.; Zhang, Q.; Sun, L.; Fan, Z. Granzyme H induces apoptosis of target tumor cells characterized by DNA fragmentation and Bid-dependent mitochondrial damage. Mol. Immunol. 2008, 45, 1044–1055. [Google Scholar] [CrossRef]
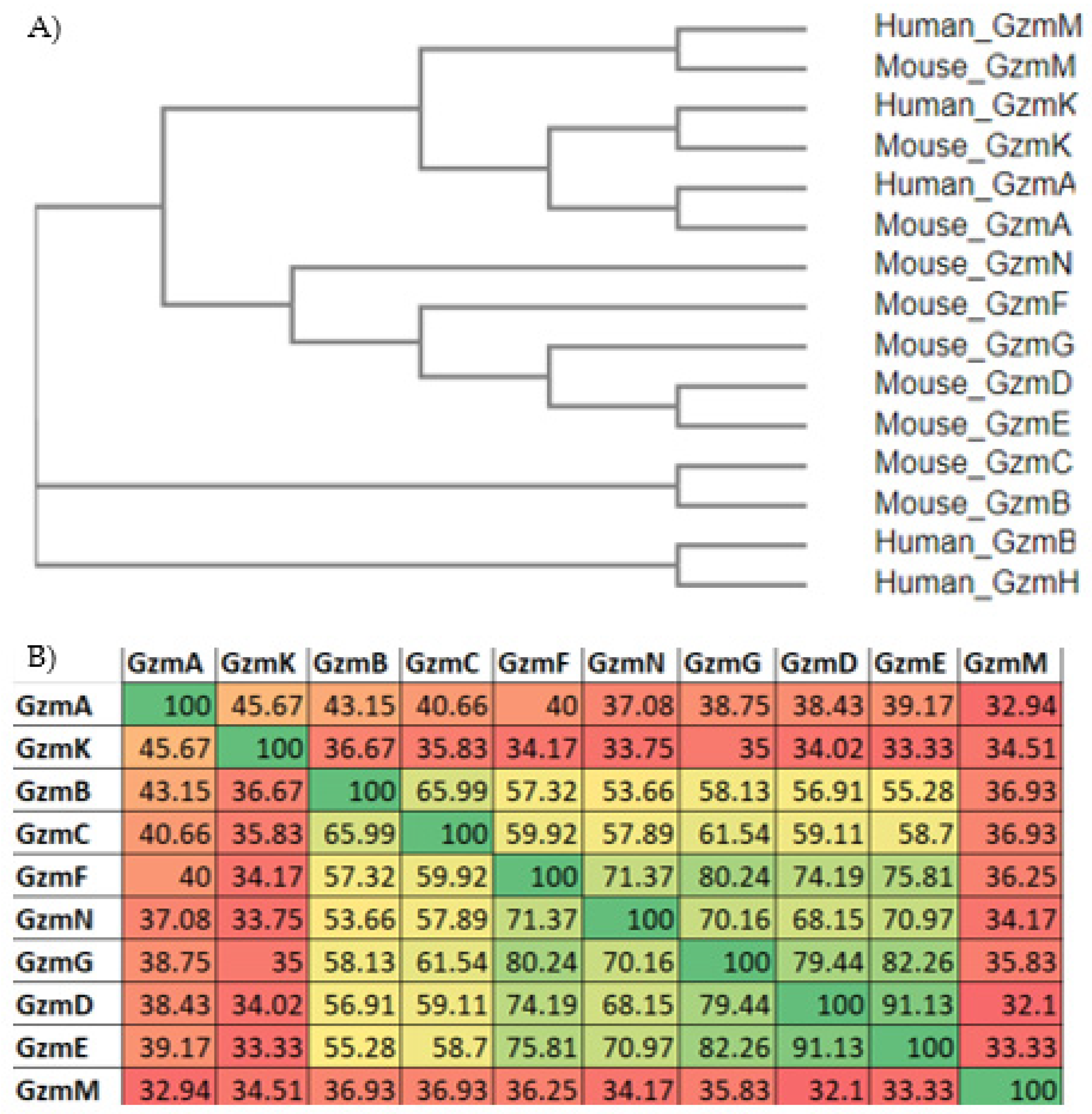


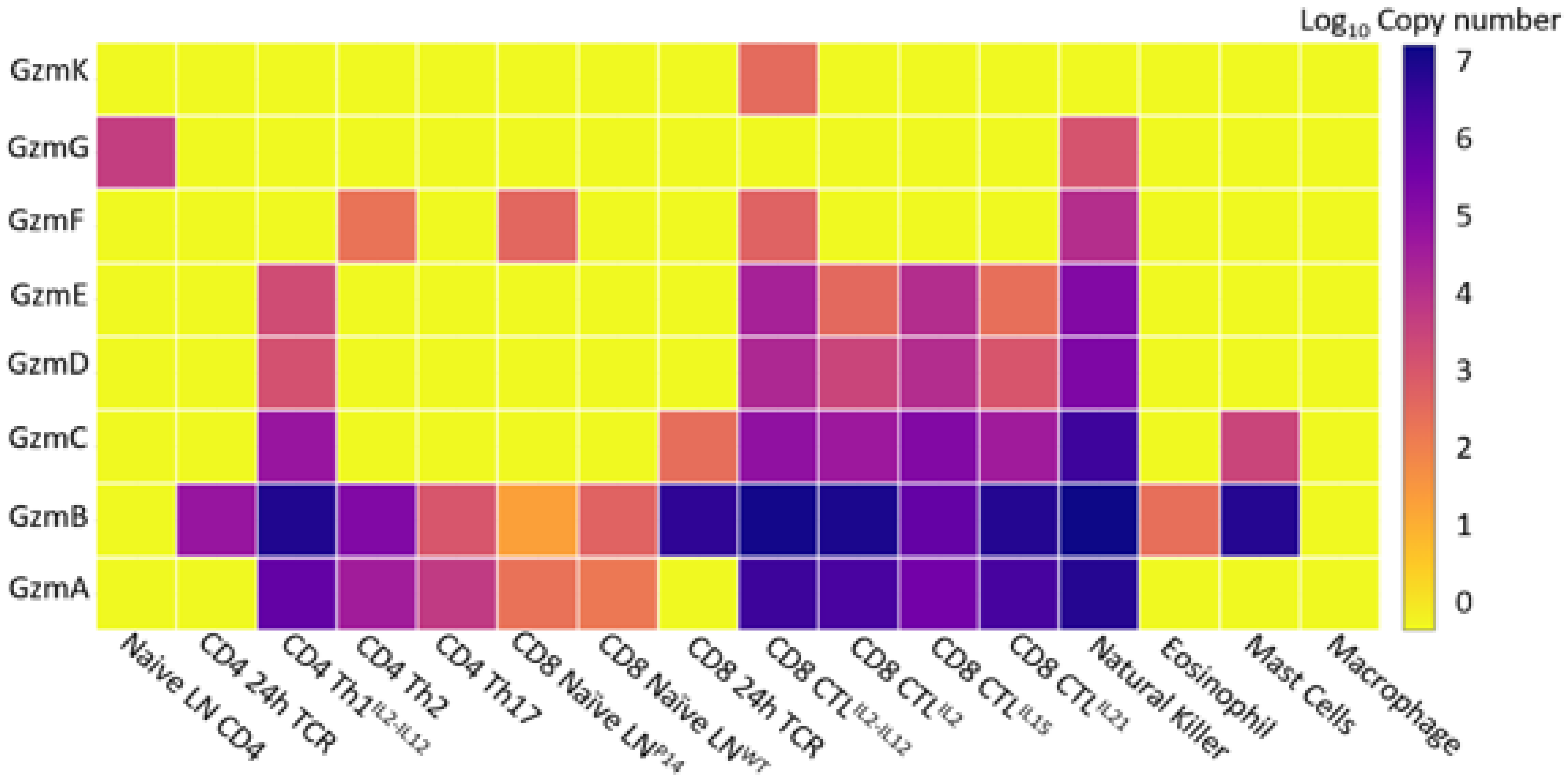
| (A) | |||
|---|---|---|---|
| Chromosome | Location | Length within Genome (bp) | |
| Gzm K | 13 c | chr13:113,308,172-113,317,431 | 9260 |
| Gzm A | 13 c | chr13:113,230,361-113,237,515 | 7155 |
| Gzm B | 14 c | chr14:56,496,295-56,499,717 | 3423 |
| Gzm C | 14 c | chr14:56,468,898-56,472,113 | 3216 |
| Gzm F | 14 c | chr14:56,442,720-56,448,874 | 6155 |
| Gzm N | 14 c | chr14:56,403,254-56,412,056 | 8803 |
| Gzm G | 14 c | chr14:56,394,039-56,397,036 | 2998 |
| Gzm D | 14 c | chr14:56,367,013-56,370,065 | 3053 |
| Gzm E | 14 c | chr14:56,355,083-56,358,082 | 3000 |
| Gzm M | 10 c | chr10:79,524,854-79,531,095 | 6242 |
| (B) | |||
| Chromosome | Location | Length within Genome (bp) | |
| Gzm K | 5 | chr5:55,024,256-55,034,570 | 10,315 |
| Gzm A | 5 | chr5:55,102,646-55,110,252 | 7607 |
| Gzm B | 14 c | chr14:24,630,954-24,634,190 | 3237 |
| Gzm H | 14 c | chr14:24,606,480-24,609,685 | 3206 |
| Gzm M | 19 c | chr19:544,053-549,922 | 5870 |
| Known Substrates | Function | |
|---|---|---|
| Gzm A * | Gasdermin B [47], SET [48,49], pro–IL-1β [50], NDUFS3 [51], histone H1 [52], HMGB2 [52], and ApeI [53] | Gzm A’s primary role is to activate and release pro-inflammatory cytokines from target cells. Mouse and human Gzm A induces pyroptosis in human cells through the cleavage and activation of human gasdermin B (there is no known mouse homologue for gasdermin B) [47]. Additionally, Gzm A can target multiple nuclear and mitochondrial targets that likely contribute to its specific form of caspase-independent cytotoxicity. |
| Gzm B * | Mouse and Human: pro-caspase 3, pro-caspase 7 Human only: Bid, inhibitor of caspase-activated DNase (ICAD), pro-caspase 8 [46] | Gzm B cell death results in the activation of the caspase cascade and induction of canonical apoptosis in both mouse and humans. |
| Gzm C | Orphan (undetermined) | Gzm C induces a caspase-independent form of cell death that results in single-stranded DNA nicking and mitochondrial swelling. Gzm C does not activate BID or the CAD nuclease, suggesting it has alternative targets and initiates different pathways from Gzm B [54]. |
| Gzm D | Orphan (undetermined) | Undetermined |
| Gzm E | Orphan (undetermined) | Undetermined |
| Gzm F | Orphan (undetermined) | Gzm F has been suggested to induce a “necroptotic-like” form of cell death[55]. Gzm F-induced cell death is caspase-independent, induces mitochondrial damage independent of the Bid pathway, results in rupture of the cellular membrane, single stranded DNA nicking, cellular organelle damage, and extensive vacuolization of the cytoplasm [55]. This death phenotype strongly resembles necroptosis [56,57]. |
| Gzm G | Orphan (undetermined) | Undetermined |
| Gzm K * | SET [58,59], β-tubulin [60], APE1 [59,61,62] | Gzm K induces single-stranded DNA damage, mitochondrial dysfunction, and generation of reactive oxygen species (ROS), and cell membrane damage through a caspase-independent mechanism [58,59]. |
| Gzm M * | Human: nucleophosmin [63], FADD [64], survivin [65], ICAD [66] Mouse and Human: alpha-Tubulin [67] | Gzm M induces apoptosis in humans though its function is undetermined in mice. Gzm M does not induce cell death in multiple mouse tumor models in vitro but mouse Gzm M can induce apoptosis in target human cancer cells in vitro [67]. |
| Gzm N | Orphan (undetermined) | To our knowledge, Gzm N has not been found to be significantly expressed in T cells or other immune cells. Its expression has been found in the testes in one study, specifically in spermatocytes and spermatids, though its biological function was not determined [68]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hay, Z.L.Z.; Slansky, J.E. Granzymes: The Molecular Executors of Immune-Mediated Cytotoxicity. Int. J. Mol. Sci. 2022, 23, 1833. https://doi.org/10.3390/ijms23031833
Hay ZLZ, Slansky JE. Granzymes: The Molecular Executors of Immune-Mediated Cytotoxicity. International Journal of Molecular Sciences. 2022; 23(3):1833. https://doi.org/10.3390/ijms23031833
Chicago/Turabian StyleHay, Zachary L. Z., and Jill E. Slansky. 2022. "Granzymes: The Molecular Executors of Immune-Mediated Cytotoxicity" International Journal of Molecular Sciences 23, no. 3: 1833. https://doi.org/10.3390/ijms23031833






