Regulation of Epstein-Barr Virus Minor Capsid Protein BORF1 by TRIM5α
Abstract
:1. Introduction
2. Results
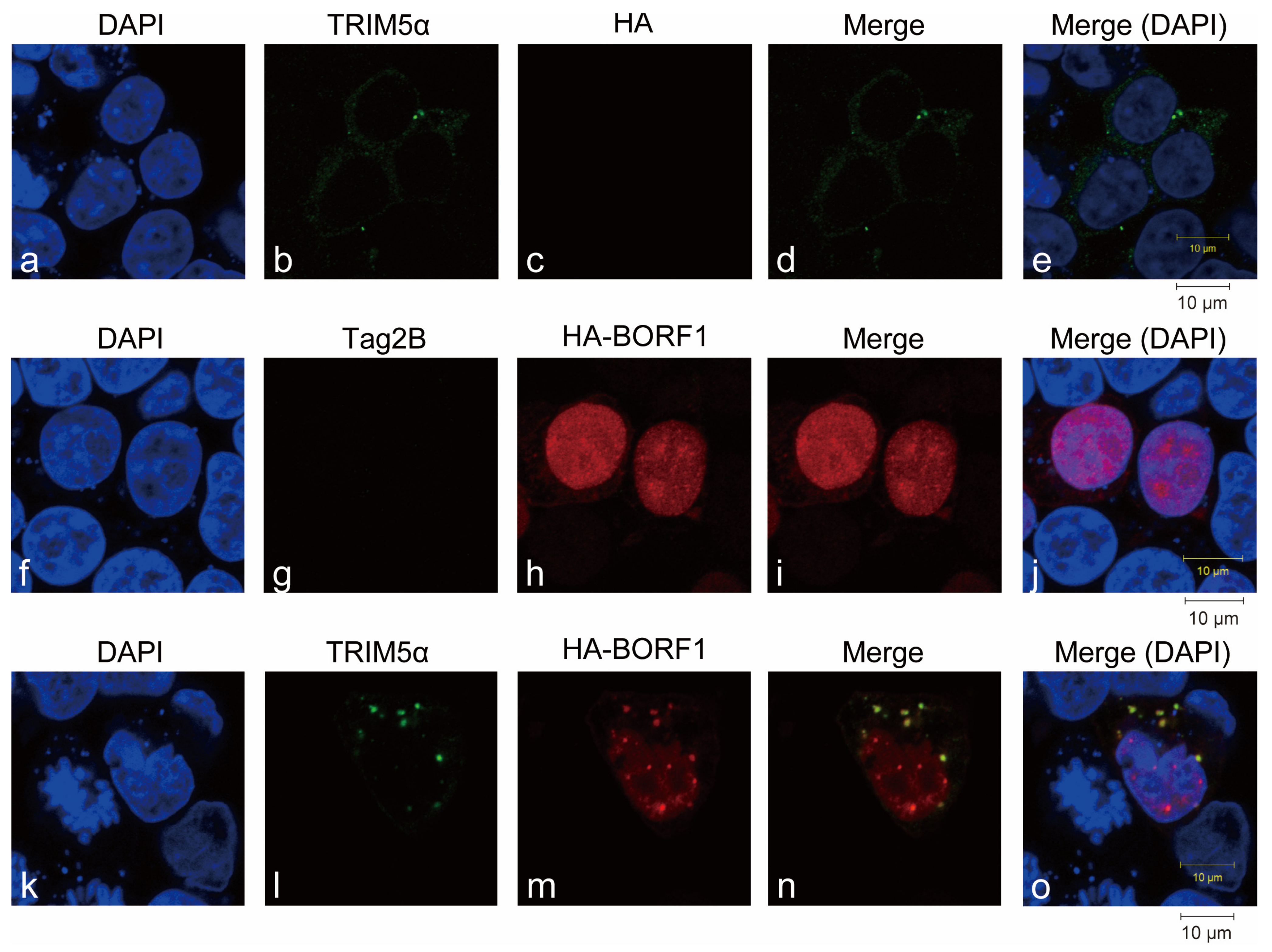
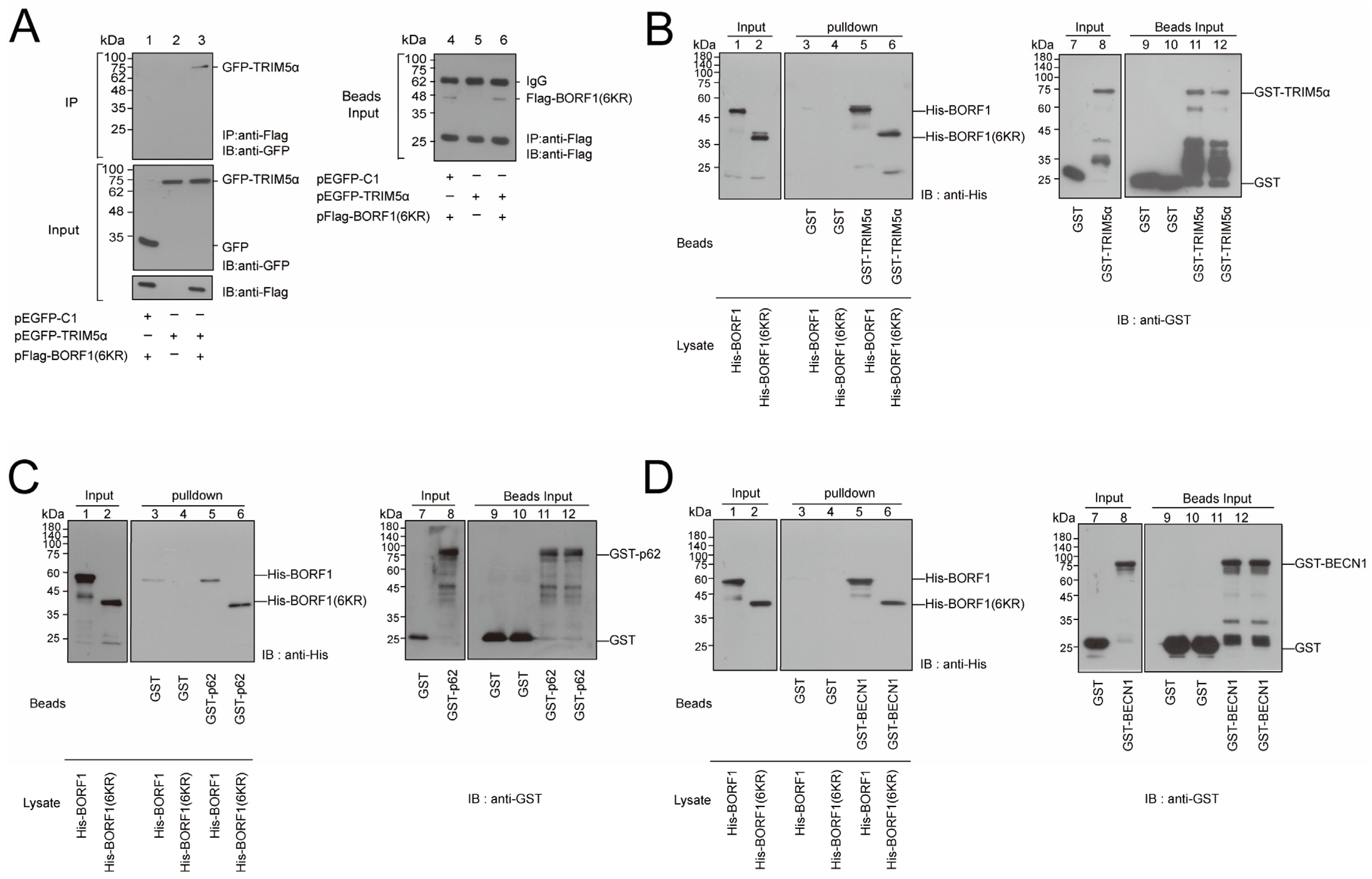
2.1. Interaction between BORF1 and TRIM5α
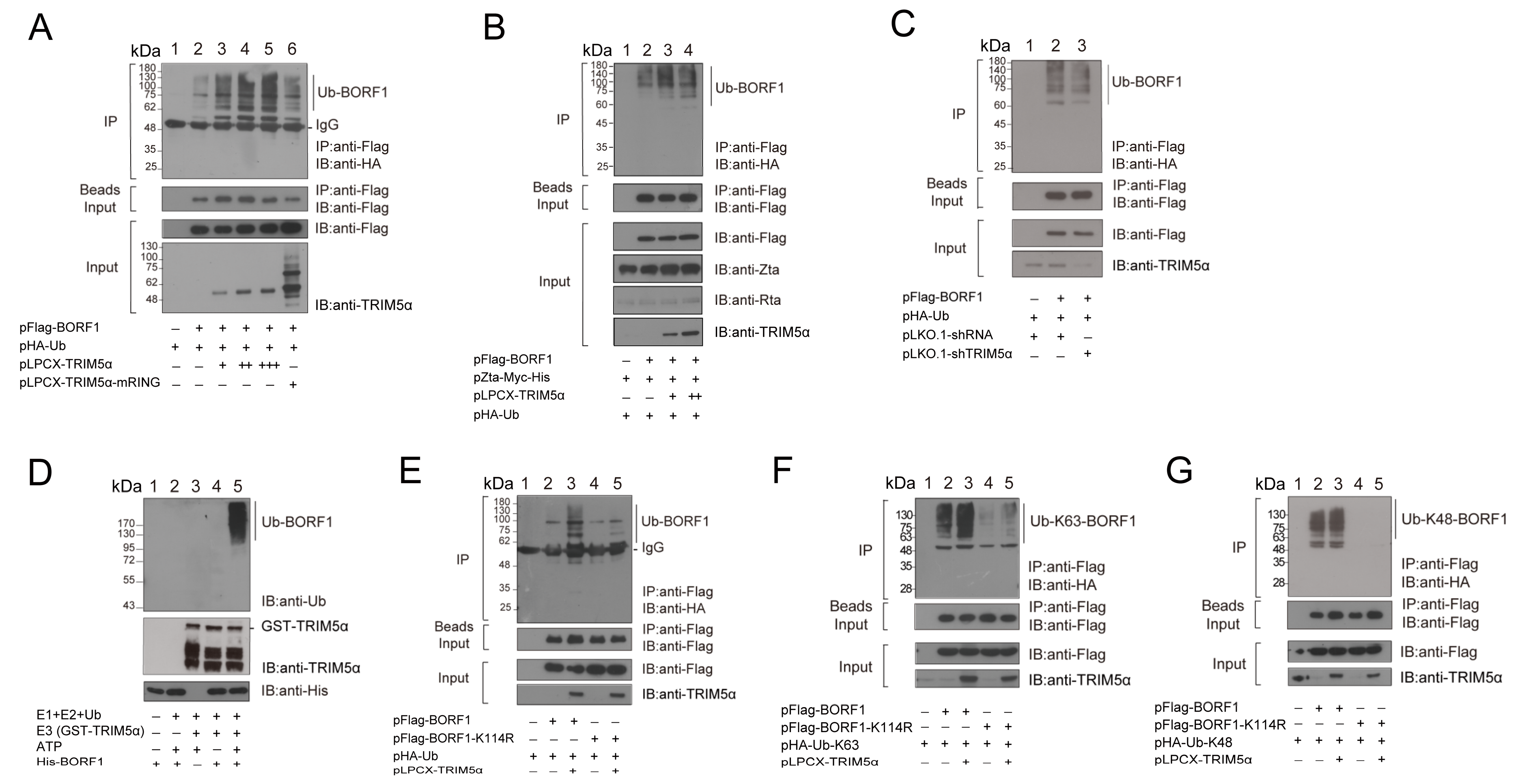
2.2. TRIM5α Is a Ubiquitin E3 Ligase of BORF1
2.3. The PRY/SPRY Domain in TRIM5α Is Critical for BORF1 Ubiquitination
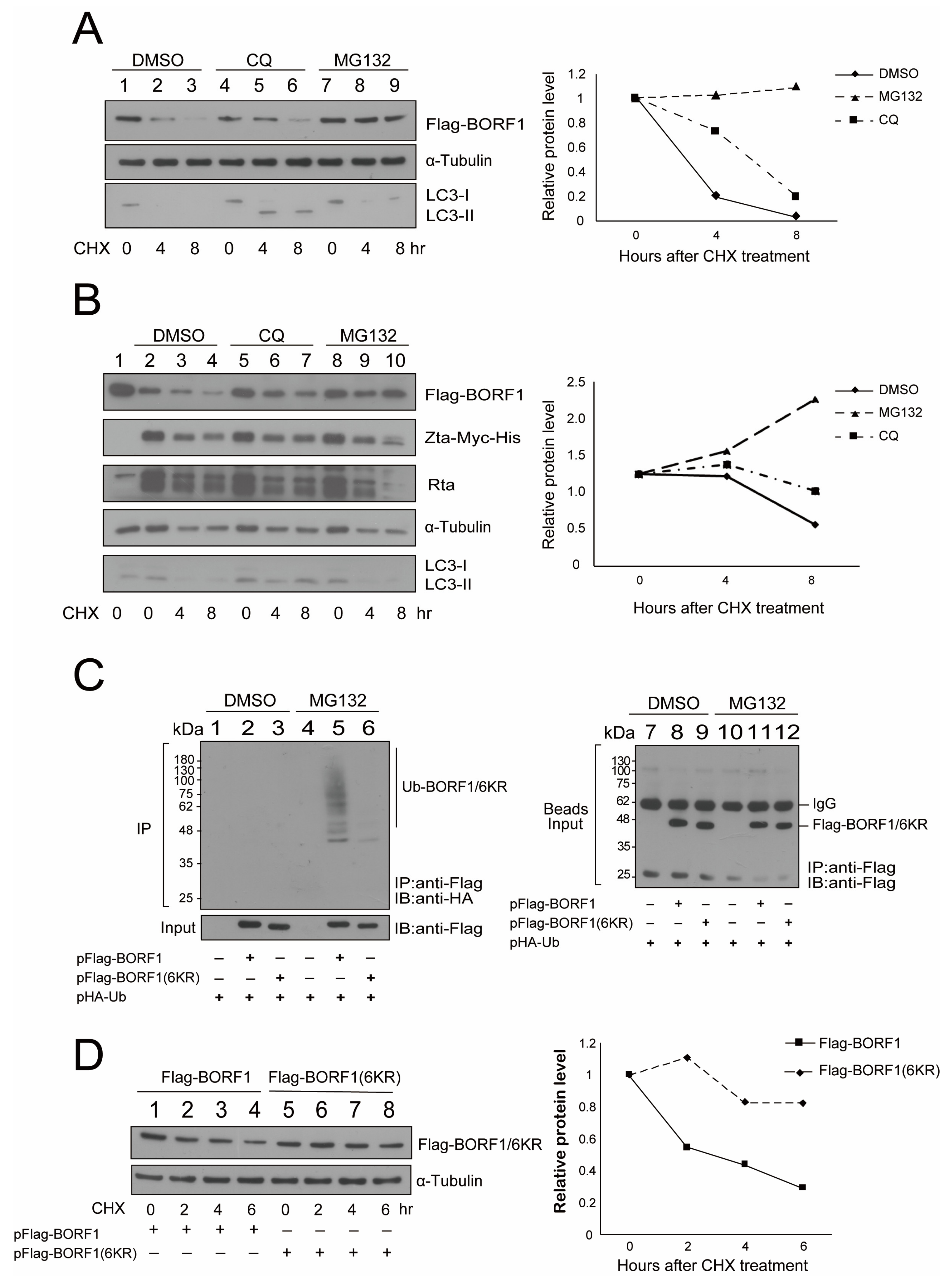
2.4. Substituting All the Lysine Residues with Arginine in BORF1 Increases Its Stability
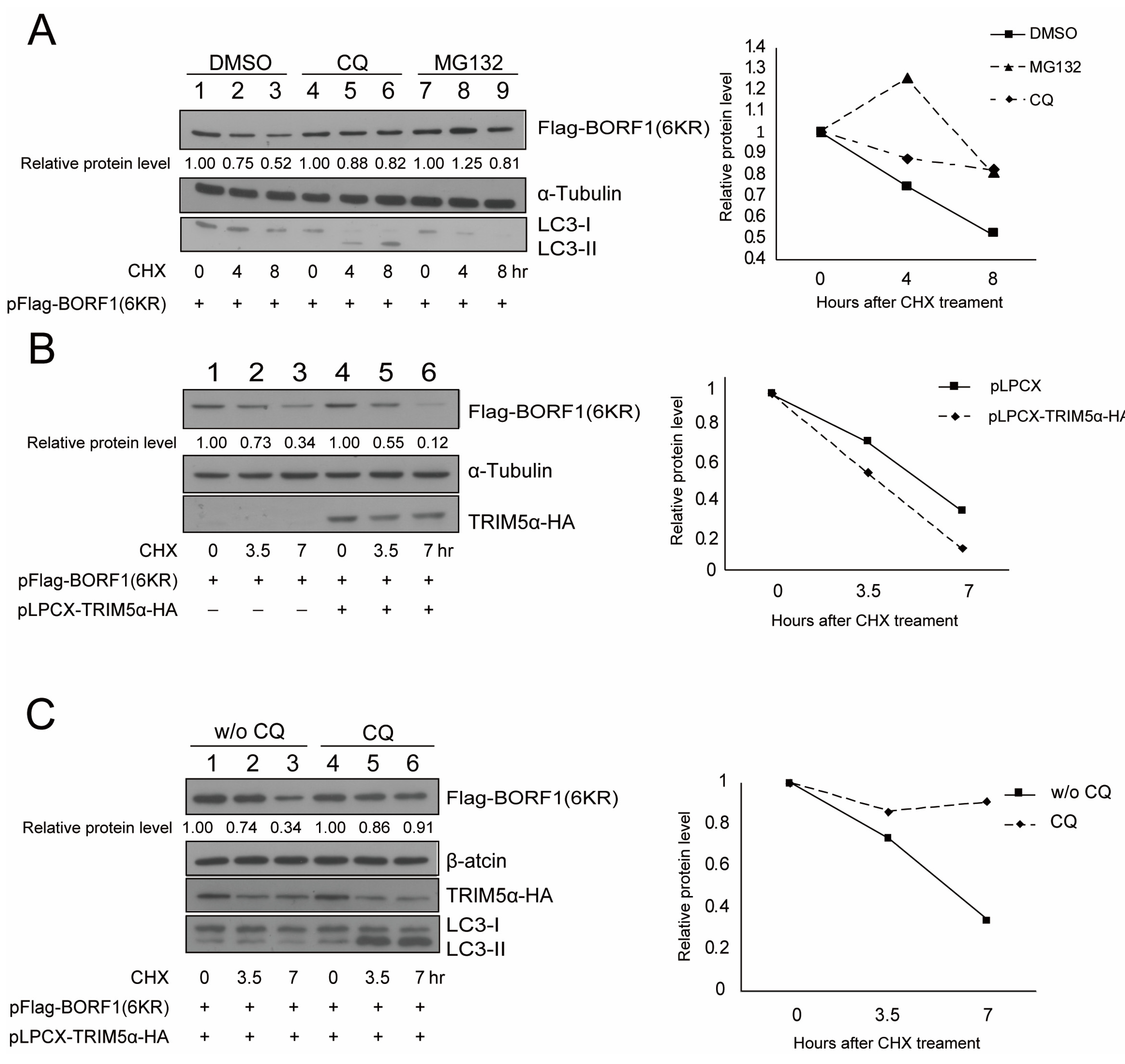
2.5. TRIM5α Destabilizes BORF1(6KR) via Autophagy
2.6. BORF1(6KR) Interacts with TRIM5α, p62, and BECN1
3. Discussion
4. Materials and Methods
4.1. Cell Line
4.2. Plasmids
4.3. Transient Transfection
4.4. Protein Production
4.5. Western Blot Analysis
4.6. Coimmunoprecipitation Assay
4.7. GST Pull-Down Analysis
4.8. Immunoprecipitation under Denaturing Conditions
4.9. In Vitro Ubiquitination Assay
4.10. Protein Stability Analysis
4.11. Immunofluorescence Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perron, M.J.; Stremlau, M.; Song, B.; Ulm, W.; Mulligan, R.C.; Sodroski, J. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11827–11832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, G.; Diez-Roux, G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 2005, 27, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Perron, M.; Welikala, S.; Sodroski, J. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 2005, 79, 3139–3145. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Griffero, F.; Li, X.; Javanbakht, H.; Song, B.; Welikala, S.; Stremlau, M.; Sodroski, J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 2006, 349, 300–315. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, K.; Wada, K.; Tanji, K.; Tanaka, M.; Kamitani, T. Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 2008, 275, 1540–1555. [Google Scholar] [CrossRef]
- Kim, J.; Tipper, C.; Sodroski, J. Role of TRIM5α RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J. Virol. 2011, 85, 8116–8132. [Google Scholar] [CrossRef] [Green Version]
- Lienlaf, M.; Hayashi, F.; Di Nunzio, F.; Tochio, N.; Kigawa, T.; Yokoyama, S.; Diaz-Griffero, F. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5α(rh): Structure of the RING domain of TRIM5α. J. Virol. 2011, 85, 8725–8737. [Google Scholar] [CrossRef] [Green Version]
- Javanbakht, H.; Yuan, W.; Yeung, D.F.; Song, B.; Diaz-Griffero, F.; Li, Y.; Li, X.; Stremlau, M.; Sodroski, J. Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 2006, 353, 234–246. [Google Scholar] [CrossRef]
- Stremlau, M.; Perron, M.; Lee, M.; Li, Y.; Song, B.; Javanbakht, H.; Diaz-Griffero, F.; Anderson, D.J.; Sundquist, W.I.; Sodroski, J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 2006, 103, 5514–5519. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Sodroski, J. The TRIM5α B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J. Virol. 2008, 82, 11495–11502. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Griffero, F.; Qin, X.R.; Hayashi, F.; Kigawa, T.; Finzi, A.; Sarnak, Z.; Lienlaf, M.; Yokoyama, S.; Sodroski, J. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 2009, 83, 10737–10751. [Google Scholar] [CrossRef] [Green Version]
- Perron, M.J.; Stremlau, M.; Lee, M.; Javanbakht, H.; Song, B.; Sodroski, J. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 2007, 81, 2138–2148. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.L.; Chandrasekaran, V.; Carter, S.D.; Woodward, C.L.; Christensen, D.E.; Dryden, K.A.; Pornillos, O.; Yeager, M.; Ganser-Pornillos, B.K.; Jensen, G.J.; et al. Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. Elife 2016, 5, e16269. [Google Scholar] [CrossRef]
- Fletcher, A.J.; Vaysburd, M.; Maslen, S.; Zeng, J.; Skehel, J.M.; Towers, G.J.; James, L.C. Trivalent RING Assembly on Retroviral Capsids Activates TRIM5 Ubiquitination and Innate Immune Signaling. Cell Host Microbe 2018, 24, 761–775.e6. [Google Scholar] [CrossRef] [Green Version]
- Khaminets, A.; Behl, C.; Dikic, I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016, 26, 6–16. [Google Scholar] [CrossRef]
- Mandell, M.A.; Jain, A.; Arko-Mensah, J.; Chauhan, S.; Kimura, T.; Dinkins, C.; Silvestri, G.; Münch, J.; Kirchhoff, F.; Simonsen, A.; et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 2014, 30, 394–409. [Google Scholar] [CrossRef] [Green Version]
- Mandell, M.A.; Kimura, T.; Jain, A.; Johansen, T.; Deretic, V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy 2014, 10, 2387–2388. [Google Scholar] [CrossRef]
- Di Rienzo, M.; Romagnoli, A.; Antonioli, M.; Piacentini, M.; Fimia, G.M. TRIM proteins in autophagy: Selective sensors in cell damage and innate immune responses. Cell Death Differ. 2020, 27, 887–902. [Google Scholar] [CrossRef]
- Kimura, T.; Mandell, M.; Deretic, V. Precision autophagy directed by receptor regulators-emerging examples within the TRIM family. J. Cell Sci. 2016, 129, 881–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reszka, N.; Zhou, C.; Song, B.; Sodroski, J.G.; Knipe, D.M. Simian TRIM5α proteins reduce replication of herpes simplex virus. Virology 2010, 398, 243–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiramel, A.I.; Meyerson, N.R.; McNally, K.L.; Broeckel, R.M.; Montoya, V.R.; Méndez-Solís, O.; Robertson, S.J.; Sturdevant, G.L.; Lubick, K.J.; Nair, V.; et al. TRIM5α Restricts Flavivirus Replication by Targeting the Viral Protease for Proteasomal Degradation. Cell Rep. 2019, 27, 3269–3283.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkmann, B.; Wittmann, S.; Lagisquet, J.; Deutschmann, J.; Eissmann, K.; Ross, J.J.; Biesinger, B.; Gramberg, T. Human TRIM5α senses and restricts LINE-1 elements. Proc. Natl. Acad. Sci. USA 2020, 117, 17965–17976. [Google Scholar] [CrossRef] [PubMed]
- Rose, K.M.; Spada, S.J.; Broeckel, R.; McNally, K.L.; Hirsch, V.M.; Best, S.M.; Bouamr, F. From Capsids to Complexes: Expanding the Role of TRIM5α in the Restriction of Divergent RNA Viruses and Elements. Viruses 2021, 13, 446. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.H.; Chen, C.S.; Wang, W.H.; Hsu, S.W.; Tsai, H.H.; Liu, S.T.; Chang, L.K. TRIM5α promotes ubiquitination of Rta from Epstein-Barr virus to attenuate lytic progression. Front. Microbiol. 2017, 7, 2129. [Google Scholar] [CrossRef] [Green Version]
- Henson, B.W.; Perkins, E.M.; Cothran, J.E.; Desai, P. Self-assembly of Epstein-Barr virus capsids. J. Virol. 2009, 83, 3877–3890. [Google Scholar] [CrossRef] [Green Version]
- Johannsen, E.; Luftig, M.; Chase, M.R.; Weicksel, S.; Cahir-McFarland, E.; Illanes, D.; Sarracino, D.; Kieff, E. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 2004, 101, 16286–16291. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.H.; Kuo, C.W.; Chang, L.K.; Hung, C.C.; Chang, T.H.; Liu, S.T. Assembly of Epstein-Barr virus capsid in promyelocytic leukemia nuclear bodies. J. Virol. 2015, 89, 8922–8931. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.H.; Chang, L.K.; Liu, S.T. Molecular interactions of Epstein-Barr virus capsid proteins. J. Virol. 2011, 85, 1615–1624. [Google Scholar] [CrossRef]
- Huang, H.H.; Wang, W.H.; Feng, T.H.; Chang, L.K. Rta is an Epstein-Barr virus tegument protein that improves the stability of capsid protein BORF1. Biochem. Biophys. Res. Commun. 2020, 523, 773–779. [Google Scholar] [CrossRef]
- Delecluse, H.J.; Hilsendegen, T.; Pich, D.; Zeidler, R.; Hammerschmidt, W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 1998, 95, 8245–8250. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Brandariz-Nunez, A.; Fricke, T.; Ivanov, D.N.; Sarnak, Z.; Diaz-Griffero, F. Binding of the rhesus TRIM5α PRYSPRY domain to capsid is necessary but not sufficient for HIV-1 restriction. Virology 2014, 448, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Hansen, M.; Rubinsztein, D.C.; Walker, D.W. Autophagy as a promoter of longevity: Insights from model organisms. Nat. Rev. Mol. Cell Biol. 2018, 19, 579–593. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Chandrasekaran, V.; Pornillos, O.; Sodroski, J.G.; Sundquist, W.I.; Yeager, M. Hexagonal assembly of a restricting TRIM5α protein. Proc. Natl. Acad. Sci. USA 2011, 108, 534–539. [Google Scholar] [CrossRef] [Green Version]
- Kocaturk, N.M.; Gozuacik, D. Crosstalk Between Mammalian Autophagy and the Ubiquitin-Proteasome System. Front. Cell Dev. Biol. 2018, 6, 128. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.H.; Chen, Y.H.; Huang, T.Y. Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 2019, 26, 80. [Google Scholar] [CrossRef]
- Wang, D.; Xu, Q.; Yuan, Q.; Jia, M.; Niu, H.; Liu, X.; Zhang, J.; Young, C.Y.; Yuan, H. Proteasome inhibition boosts autophagic degradation of ubiquitinated-AGR2 and enhances the antitumor efficiency of bevacizumab. Oncogene 2019, 38, 3458–3474. [Google Scholar] [CrossRef] [Green Version]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Chang, L.K.; Lee, Y.H.; Cheng, T.S.; Hong, Y.R.; Lu, P.J.; Wang, J.J.; Wang, W.H.; Kuo, C.W.; Li, S.S.L.; Liu, S.T. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 2004, 279, 38803–38812. [Google Scholar] [CrossRef]
- Yang, Y.C.; Yoshikai, Y.; Hsu, S.W.; Saitoh, H.; Chang, L.K. Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus. J. Biol. Chem. 2013, 288, 12866–12879. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, L.-T.; Lu, Y.-S.; Huang, H.-H.; Chen, H.; Hsu, S.-W.; Chang, L.-K. Regulation of Epstein-Barr Virus Minor Capsid Protein BORF1 by TRIM5α. Int. J. Mol. Sci. 2022, 23, 15340. https://doi.org/10.3390/ijms232315340
Lin L-T, Lu Y-S, Huang H-H, Chen H, Hsu S-W, Chang L-K. Regulation of Epstein-Barr Virus Minor Capsid Protein BORF1 by TRIM5α. International Journal of Molecular Sciences. 2022; 23(23):15340. https://doi.org/10.3390/ijms232315340
Chicago/Turabian StyleLin, Lih-Tsern, Yi-Shan Lu, Hsiang-Hung Huang, Hao Chen, Shih-Wei Hsu, and Li-Kwan Chang. 2022. "Regulation of Epstein-Barr Virus Minor Capsid Protein BORF1 by TRIM5α" International Journal of Molecular Sciences 23, no. 23: 15340. https://doi.org/10.3390/ijms232315340



