Ferulic Acid and P-Coumaric Acid Synergistically Attenuate Non-Alcoholic Fatty Liver Disease through HDAC1/PPARG-Mediated Free Fatty Acid Uptake
Abstract
:1. Introduction
2. Results
2.1. Prediction of the Association of FA/p-CA with Non-alcoholic Fatty Liver Disease
2.2. FA/p-CA Treatment Suppresses FFA-Induced Lipid Accumulation In Vitro
2.3. FA/p-CA Attenuate HFD-Induced Hepatic Injury and Steatosis In Vivo
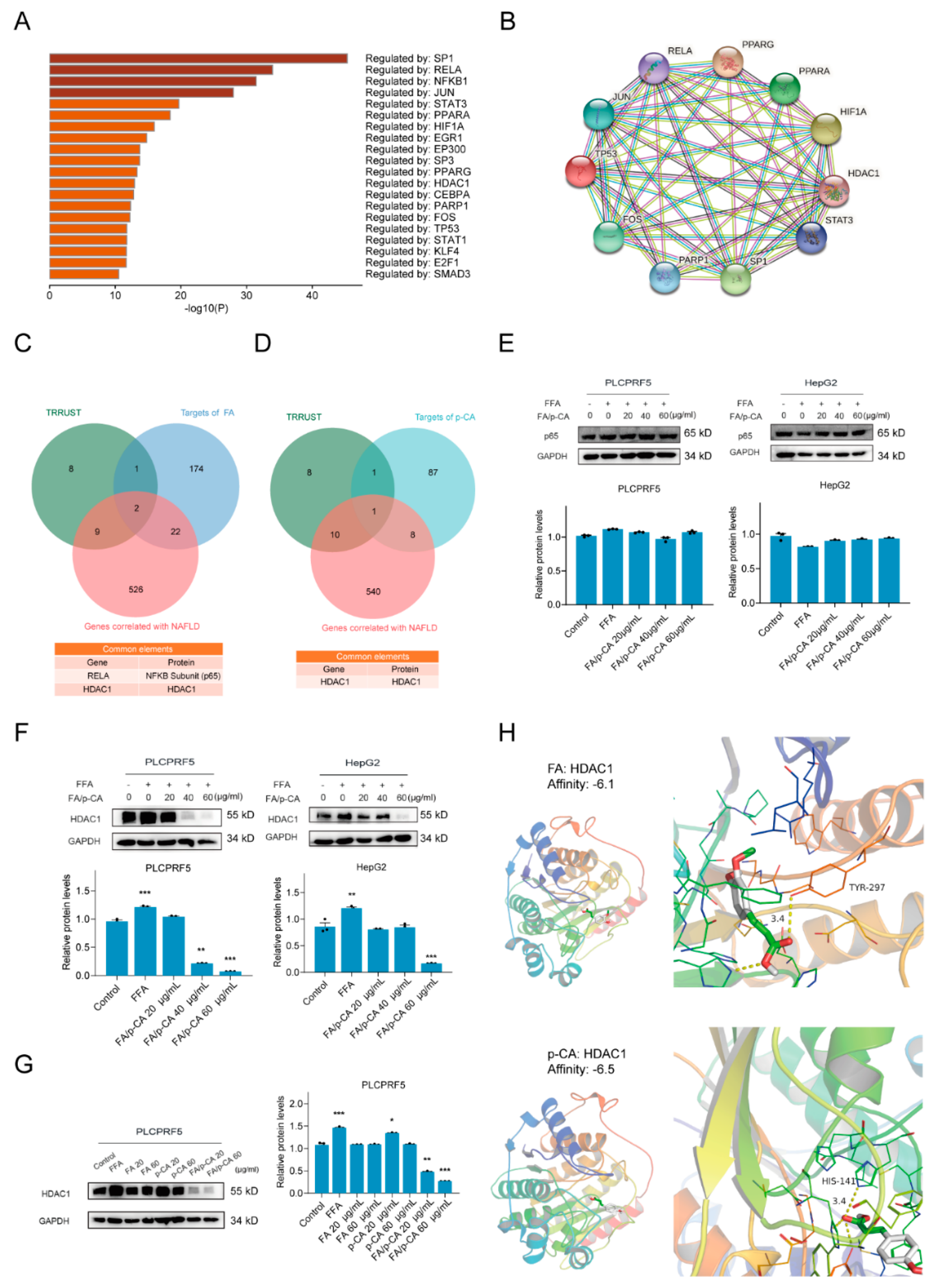
2.4. The Potential Pathways of FA p-CA Targets
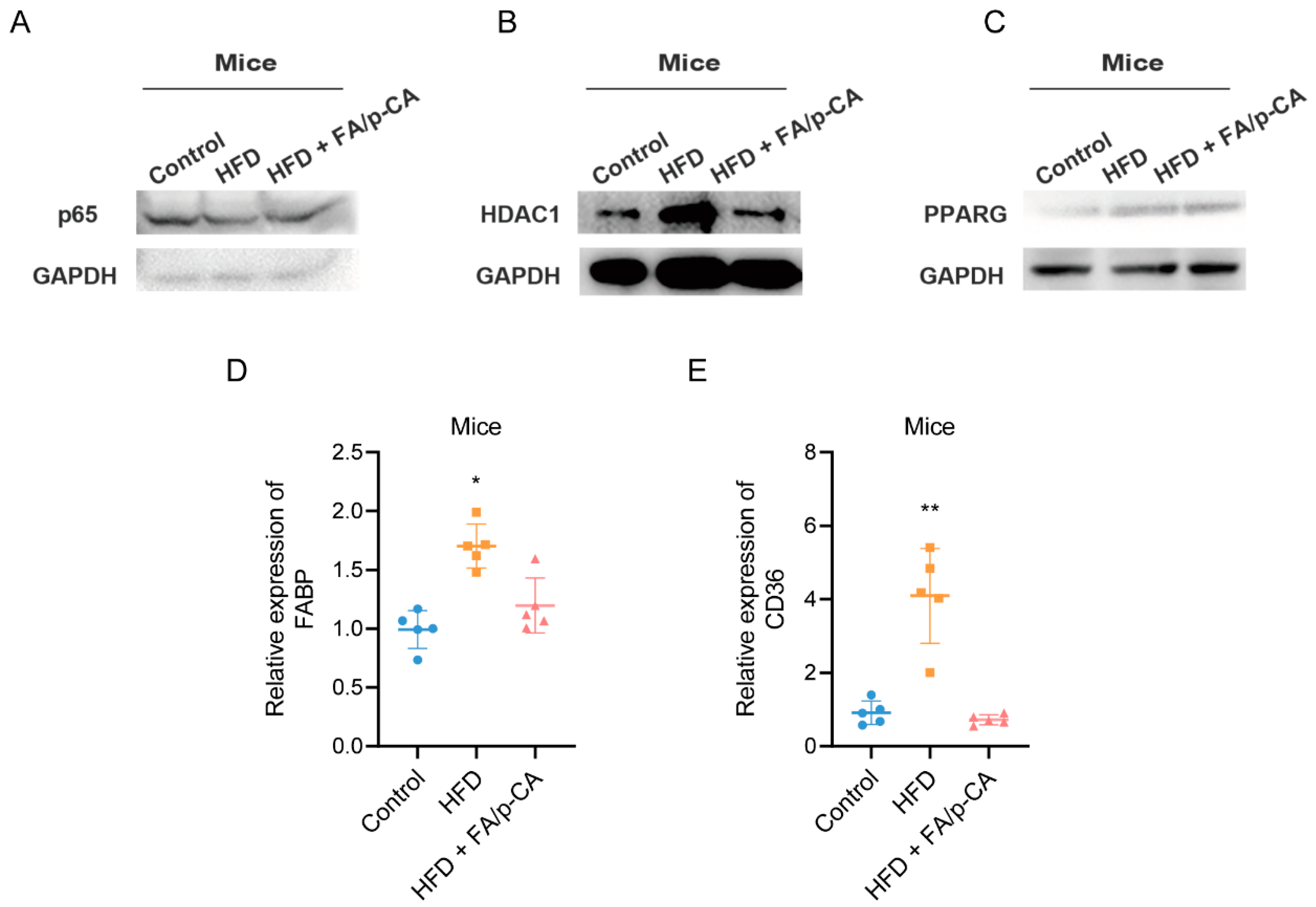
2.5. HDAC1 Is Involved in Amelioration of NAFLD by FA/p-CA
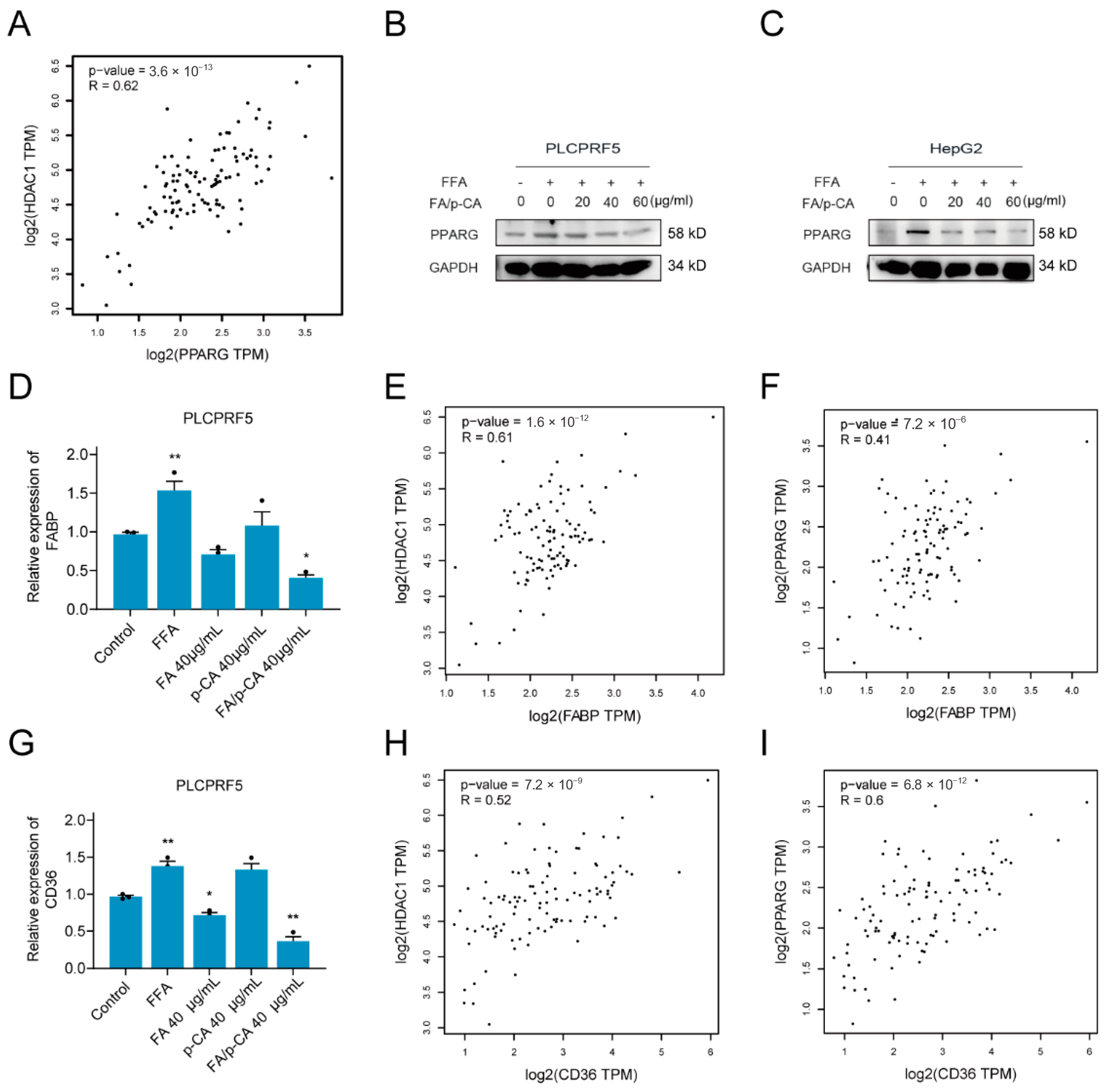
2.6. FA/p-CA Inhibit Hepatic Lipid Uptake Via the HDAC1/PPARG Axis
3. Discussion
4. Martials and Methods
4.1. Materials
4.2. Cell Culture and Steatosis Induction
4.3. Bodipy and Oil Red O Staining
4.4. Triglyceride Assay
4.5. Mice Experiment
4.6. Histological Examinations
4.7. Western Blot
4.8. Quantitative RT-PCR
4.9. Molecular Docking
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rinella, M.E. Nonalcoholic fatty liver disease: A systematic review. Jama 2015, 313, 2263–2273. [Google Scholar] [CrossRef]
- Draijer, L.; Benninga, M.; Koot, B. Pediatric NAFLD: An overview and recent developments in diagnostics and treatment. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 447–461. [Google Scholar] [CrossRef]
- Sheka, A.C.; Adeyi, O.; Thompson, J.; Hameed, B.; Crawford, P.A.; Ikramuddin, S. Nonalcoholic Steatohepatitis: A Review. Jama 2020, 323, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.J.; Aguilar, M.; Cheung, R.; Perumpail, R.B.; Harrison, S.A.; Younossi, Z.M.; Ahmed, A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015, 148, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.C.; Horton, J.D.; Hobbs, H.H. Human fatty liver disease: Old questions and new insights. Science (N. Y.) 2011, 332, 1519–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuppan, D.; Gorrell, M.D.; Klein, T.; Mark, M.; Afdhal, N.H. The challenge of developing novel pharmacological therapies for non-alcoholic steatohepatitis. Liver Int. Off. J. Int. Assoc. Study Liver 2010, 30, 795–808. [Google Scholar] [CrossRef]
- Lazarus, J.V.; Mark, H.E.; Villota-Rivas, M.; Palayew, A.; Carrieri, P.; Colombo, M.; Ekstedt, M.; Esmat, G.; George, J.; Marchesini, G.; et al. The global NAFLD policy review and preparedness index: Are countries ready to address this silent public health challenge? J. Hepatol. 2022, 76, 771–780. [Google Scholar] [CrossRef]
- Boccellino, M.; D’Angelo, S. Anti-Obesity Effects of Polyphenol Intake: Current Status and Future Possibilities. Int. J. Mol. Sci. 2020, 21, 5642. [Google Scholar] [CrossRef]
- Sireesha, Y.; Kasetti, R.B.; Nabi, S.A.; Swapna, S.; Apparao, C. Antihyperglycemic and hypolipidemic activities of Setaria italica seeds in STZ diabetic rats. Pathophysiology 2011, 18, 159–164. [Google Scholar] [CrossRef]
- Lu, Y.; Shan, S.; Li, H.; Shi, J.; Zhang, X.; Li, Z. Reversal Effects of Bound Polyphenol from Foxtail Millet Bran on Multidrug Resistance in Human HCT-8/Fu Colorectal Cancer Cell. J. Agric. Food Chem. 2018, 66, 5190–5199. [Google Scholar] [CrossRef]
- Ou, Q.; Zhang, S.; Fu, C.; Yu, L.; Xin, P.; Gu, Z.; Cao, Z.; Wu, J.; Wang, Y. More natural more better: Triple natural anti-oxidant puerarin/ferulic acid/polydopamine incorporated hydrogel for wound healing. J. Nanobiotechnol. 2021, 19, 237. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Shen, J.D.; Xu, L.P.; Li, H.B.; Li, Y.C.; Yi, L.T. Ferulic acid inhibits neuro-inflammation in mice exposed to chronic unpredictable mild stress. Int. Immunopharmacol. 2017, 45, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Mu, M.; Zuo, S.; Wu, R.M.; Deng, K.S.; Lu, S.; Zhu, J.J.; Zou, G.L.; Yang, J.; Cheng, M.L.; Zhao, X.K. Ferulic acid attenuates liver fibrosis and hepatic stellate cell activation via inhibition of TGF-β/Smad signaling pathway. Drug Des. Dev. Ther. 2018, 12, 4107–4115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, K.; Ou, J.; Huang, J.; Ou, S. p-Coumaric acid and its conjugates: Dietary sources, pharmacokinetic properties and biological activities. J. Sci. Food Agric. 2016, 96, 2952–2962. [Google Scholar] [CrossRef]
- de Anda-Jáuregui, G.; McGregor, B.A.; Guo, K.; Hur, J. A Network Pharmacology Approach for the Identification of Common Mechanisms of Drug-Induced Peripheral Neuropathy. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Jacunski, A.; Tatonetti, N.P. Connecting the dots: Applications of network medicine in pharmacology and disease. Clin. Pharmacol. Ther. 2013, 94, 659–669. [Google Scholar] [CrossRef]
- Petroni, M.L.; Brodosi, L.; Barbanti, F.A.; di Domizio, S.; Petta, S.; Marchesini, G. Lifestyle Changes for the Treatment of Nonalcoholic Fatty Liver Disease—A 2015-19 Update. Curr. Pharm. Des. 2020, 26, 1110–1118. [Google Scholar] [CrossRef]
- Diehl, A.M.; Day, C. Cause, Pathogenesis, and Treatment of Nonalcoholic Steatohepatitis. New Engl. J. Med. 2017, 377, 2063–2072. [Google Scholar] [CrossRef]
- Wang, B.H.; Ou-Yang, J.P. Pharmacological actions of sodium ferulate in cardiovascular system. Cardiovasc. Drug Rev. 2005, 23, 161–172. [Google Scholar] [CrossRef]
- Liu, F.; Shan, S.; Li, H.; Shi, J.; Hao, R.; Yang, R.; Li, Z. Millet shell polyphenols prevent atherosclerosis by protecting the gut barrier and remodeling the gut microbiota in ApoE(-/-) mice. Food Funct. 2021, 12, 7298–7309. [Google Scholar] [CrossRef]
- Ed Nignpense, B.; Francis, N.; Blanchard, C.; Santhakumar, A.B. Bioaccessibility and Bioactivity of Cereal Polyphenols: A Review. Foods 2021, 10, 1595. [Google Scholar] [CrossRef] [PubMed]
- Awika, J.M.; Rose, D.J.; Simsek, S. Complementary effects of cereal and pulse polyphenols and dietary fiber on chronic inflammation and gut health. Food Funct. 2018, 9, 1389–1409. [Google Scholar] [CrossRef]
- Jahn, D.; Kircher, S.; Hermanns, H.M.; Geier, A. Animal models of NAFLD from a hepatologist’s point of view. Biochim. Biophys. Acta. Mol. Basis Dis. 2019, 1865, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Recena Aydos, L.; Aparecida do Amaral, L.; Serafim de Souza, R.; Jacobowski, A.C.; Freitas Dos Santos, E.; Rodrigues Macedo, M.L. Nonalcoholic Fatty Liver Disease Induced by High-Fat Diet in C57bl/6 Models. Nutrients 2019, 11, 3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanji, A.A. Animal models of nonalcoholic fatty liver disease and steatohepatitis. Clin. Liver Dis. 2004, 8, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.M.; Kwon, Y.E.; Kim, J.M.; Bae, S.J.; Lee, B.K.; Yoo, S.J.; Chung, C.H.; Deshaies, R.J.; Seol, J.H. Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat. Cell Biol. 2009, 11, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Liu, X.; Chen, J.; Gao, S.; Lu, L.; Zhang, H.; Ding, G.; Wang, Z.; Chen, Z.; Shi, T.; et al. Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017, 27, 898–915. [Google Scholar] [CrossRef] [Green Version]
- Kawano, Y.; Cohen, D.E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J. Gastroenterol. 2013, 48, 434–441. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Zhang, X.; Zhao, Z.; Lu, H.; Ke, B.; Ye, X.; Wu, B.; Ye, J. NF-κB/HDAC1/SREBP1c pathway mediates the inflammation signal in progression of hepatic steatosis. Acta Pharm. Sin. B 2020, 10, 825–836. [Google Scholar] [CrossRef]
- Lai, P.H.; Wang, W.L.; Ko, C.Y.; Lee, Y.C.; Yang, W.M.; Shen, T.W.; Chang, W.C.; Wang, J.M. HDAC1/HDAC3 modulates PPARG2 transcription through the sumoylated CEBPD in hepatic lipogenesis. Biochim. Biophys. Acta 2008, 1783, 1803–1814. [Google Scholar] [CrossRef]
- Hao, J.W.; Wang, J.; Guo, H.; Zhao, Y.Y.; Sun, H.H.; Li, Y.F.; Lai, X.Y.; Zhao, N.; Wang, X.; Xie, C.; et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 2020, 11, 4765. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, R.W.; Holloway, G.P.; Luiken, J.J.; Bonen, A.; Glatz, J.F. Fatty acid transport across the cell membrane: Regulation by fatty acid transporters. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wan, M.; Xu, D.; Pan, D.; Xia, H.; Yang, L.; Sun, G. Flaxseed Powder Attenuates Non-Alcoholic Steatohepatitis via Modulation of Gut Microbiota and Bile Acid Metabolism through Gut-Liver Axis. Int. J. Mol. Sci. 2021, 22, 10858. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Gao, G.; Wang, H.; Li, E.; Yuan, Y.; Xu, J.; Zhang, Z.; Wang, P.; Fu, Y.; Zeng, H.; et al. Dehydroabietic acid alleviates high fat diet-induced insulin resistance and hepatic steatosis through dual activation of PPAR-γ and PPAR-α. Biomed. Pharmacother. 2020, 127, 110155. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
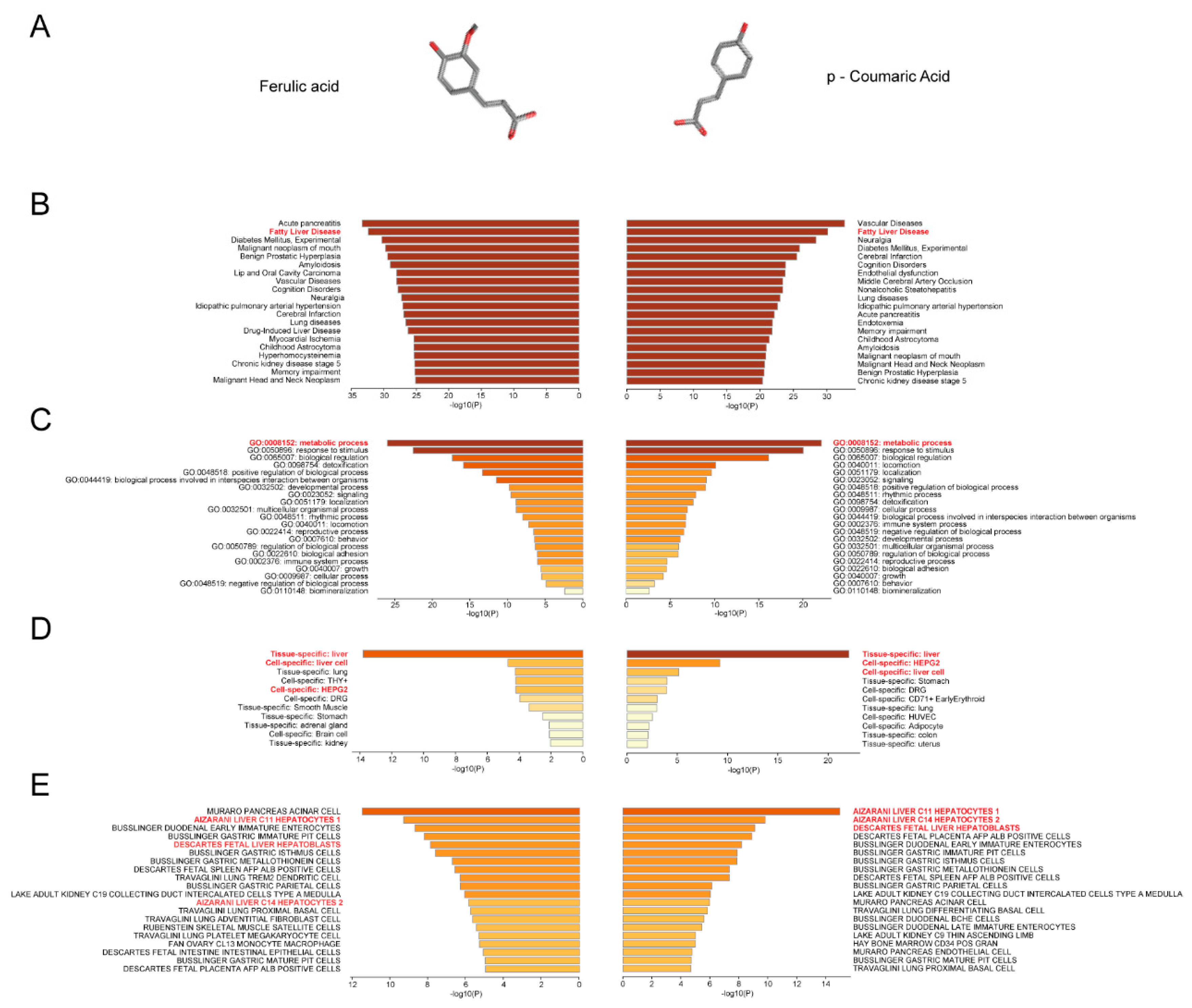


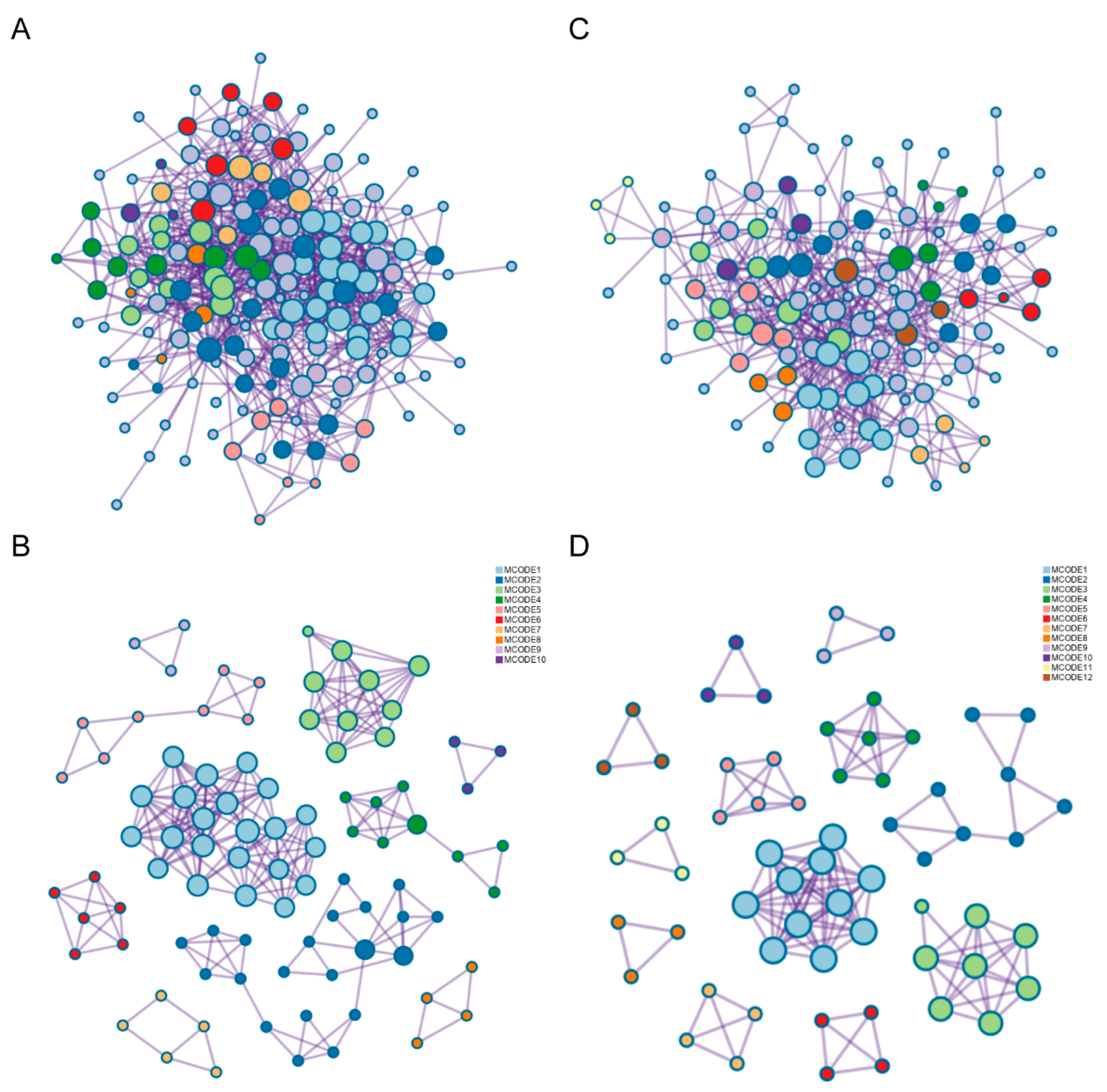
| MCODE | GO | Description | Log10(P) |
|---|---|---|---|
| MCODE_1 | R-HSA-190840 | Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane | −14.9 |
| R-HSA-190872 | Transport of connexons to the plasma membrane | −14.7 | |
| R-HSA-389977 | Post-chaperonin tubulin folding pathway | −14.3 | |
| MCODE_2 | GO:1901615 | organic hydroxy compound metabolic process | −12.7 |
| hsa00350 | Tyrosine metabolism | −10.4 | |
| GO:0010817 | regulation of hormone levels | −9.1 | |
| MCODE_3 | M207 | PID RETINOIC ACID PATHWAY | −19.3 |
| R-HSA-383280 | Nuclear Receptor transcription pathway | −17.5 | |
| M162 | PID RXR VDR PATHWAY | −16.3 | |
| MCODE_4 | WP167 | Eicosanoid synthesis | −16.6 |
| WP5122 | Prostaglandin and leukotriene metabolism in senescence | −16.2 | |
| GO:0019369 | arachidonic acid metabolic process | −14.5 | |
| MCODE_5 | R-HSA-211859 | Biological oxidations | −17.1 |
| WP702 | Metapathway biotransformation Phase I and II | −14.6 | |
| R-HSA-156580 | Phase II—Conjugation of compounds | −13.3 | |
| MCODE_6 | hsa05130 | Pathogenic Escherichia coli infection | −13.1 |
| hsa05417 | Lipid and atherosclerosis | −12.9 | |
| hsa05131 | Shigellosis | −12.6 | |
| MCODE_7 | M186 | PID PDGFRB PATHWAY | −6.1 |
| R-HSA-1280215 | Cytokine Signaling in Immune system | −5.8 | |
| GO:0030100 | regulation of endocytosis | −5.5 | |
| MCODE_8 | WP272 | Blood clotting cascade | −8.8 |
| WP558 | Complement and coagulation cascades | −7.5 | |
| GO:0030193 | regulation of blood coagulation | −7.4 | |
| MCODE_9 | WP702 | Metapathway biotransformation Phase I and II | −6.6 |
| MCODE | GO | Description | Log10(P) |
|---|---|---|---|
| MCODE_1 | R-HSA-2122947 | NOTCH1 Intracellular Domain Regulates Transcription | −27.4 |
| R-HSA-2894858 | Signaling by NOTCH1 HD+PEST Domain Mutants in Cancer | −26.5 | |
| R-HSA-2894862 | Constitutive Signaling by NOTCH1 HD+PEST Domain Mutants | −26.5 | |
| MCODE_2 | R-HSA-1592389 | Activation of Matrix Metalloproteinases | −7 |
| M174 | PID UPA UPAR PATHWAY | −6.7 | |
| WP534 | Glycolysis and gluconeogenesis | −6.6 | |
| MCODE_3 | GO:0046394 | carboxylic acid biosynthetic process | −6.4 |
| GO:0016053 | organic acid biosynthetic process | −6.4 | |
| GO:1901607 | alpha-amino acid biosynthetic process | −6.2 | |
| MCODE_4 | R-HSA-418594 | G alpha (i) signaling events | −11.9 |
| R-HSA-373076 | Class A/1 (Rhodopsin-like receptors) | −11.8 | |
| R-HSA-500792 | GPCR ligand binding | −10.9 | |
| MCODE_6 | R-HSA-6798695 | Neutrophil degranulation | −7.2 |
| R-HSA-1474244 | Extracellular matrix organization | −5.4 | |
| MCODE_7 | hsa00982 | Drug metabolism—cytochrome P450 | −10.5 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | −10.4 | |
| R-HSA-211859 | Biological oxidations | −8.5 | |
| MCODE_8 | R-HSA-383280 | Nuclear receptor transcription pathway | −8.3 |
| MCODE_9 | WP702 | Metapathway biotransformation Phase I and II | −6.6 |
| MCODE_11 | R-HSA-6798695 | Neutrophil degranulation | −5.4 |
| Genes | Sequences (5′-3′) | |
|---|---|---|
| FABP | forward primer: | reverse primer: |
| TGGCGTTTGACAGCACTTGG | AGCTTCAAATTGTCATGAGCTGCA | |
| CD36 | forward primer: | reverse primer: |
| TCTCAATCTGGCTGTGGCAG | CAGGGTACGGAACCAAACTCA | |
| GAPDH | forward primer: | reverse primer: |
| GCACCGTCAAGGCTGAGAAC | TGGTGAAGAACGCCAGTGGA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, K.; Zhang, L.; La, X.; Wu, H.; Yang, R.; Li, H.; Li, Z. Ferulic Acid and P-Coumaric Acid Synergistically Attenuate Non-Alcoholic Fatty Liver Disease through HDAC1/PPARG-Mediated Free Fatty Acid Uptake. Int. J. Mol. Sci. 2022, 23, 15297. https://doi.org/10.3390/ijms232315297
Cui K, Zhang L, La X, Wu H, Yang R, Li H, Li Z. Ferulic Acid and P-Coumaric Acid Synergistically Attenuate Non-Alcoholic Fatty Liver Disease through HDAC1/PPARG-Mediated Free Fatty Acid Uptake. International Journal of Molecular Sciences. 2022; 23(23):15297. https://doi.org/10.3390/ijms232315297
Chicago/Turabian StyleCui, Kaili, Lichao Zhang, Xiaoqin La, Haili Wu, Ruipeng Yang, Hanqing Li, and Zhuoyu Li. 2022. "Ferulic Acid and P-Coumaric Acid Synergistically Attenuate Non-Alcoholic Fatty Liver Disease through HDAC1/PPARG-Mediated Free Fatty Acid Uptake" International Journal of Molecular Sciences 23, no. 23: 15297. https://doi.org/10.3390/ijms232315297




