In Vitro Anti-Proliferative and Apoptotic Effects of Hydroxytyrosyl Oleate on SH-SY5Y Human Neuroblastoma Cells
Abstract
:1. Introduction
2. Results
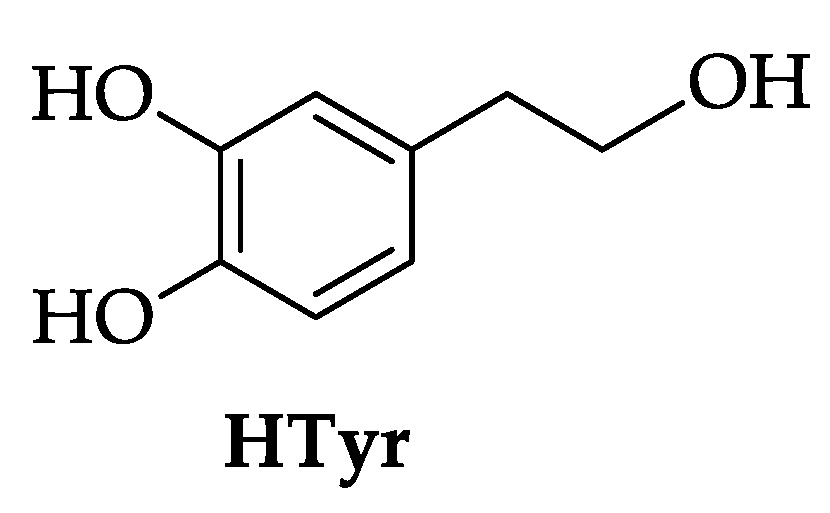
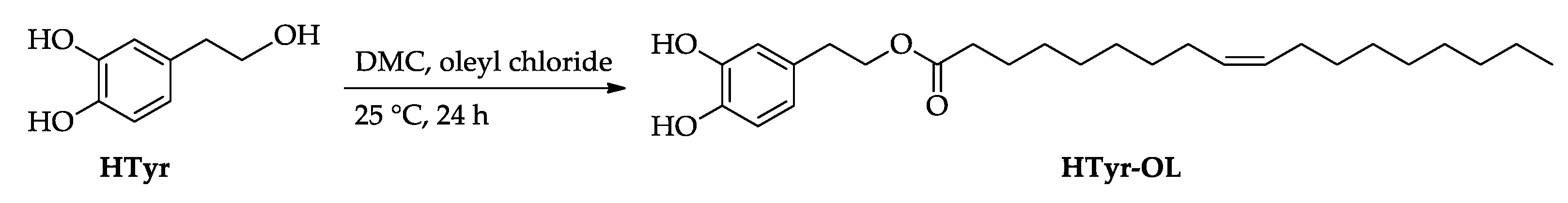
2.1. Synthesis of HTyr and HTyr-OL
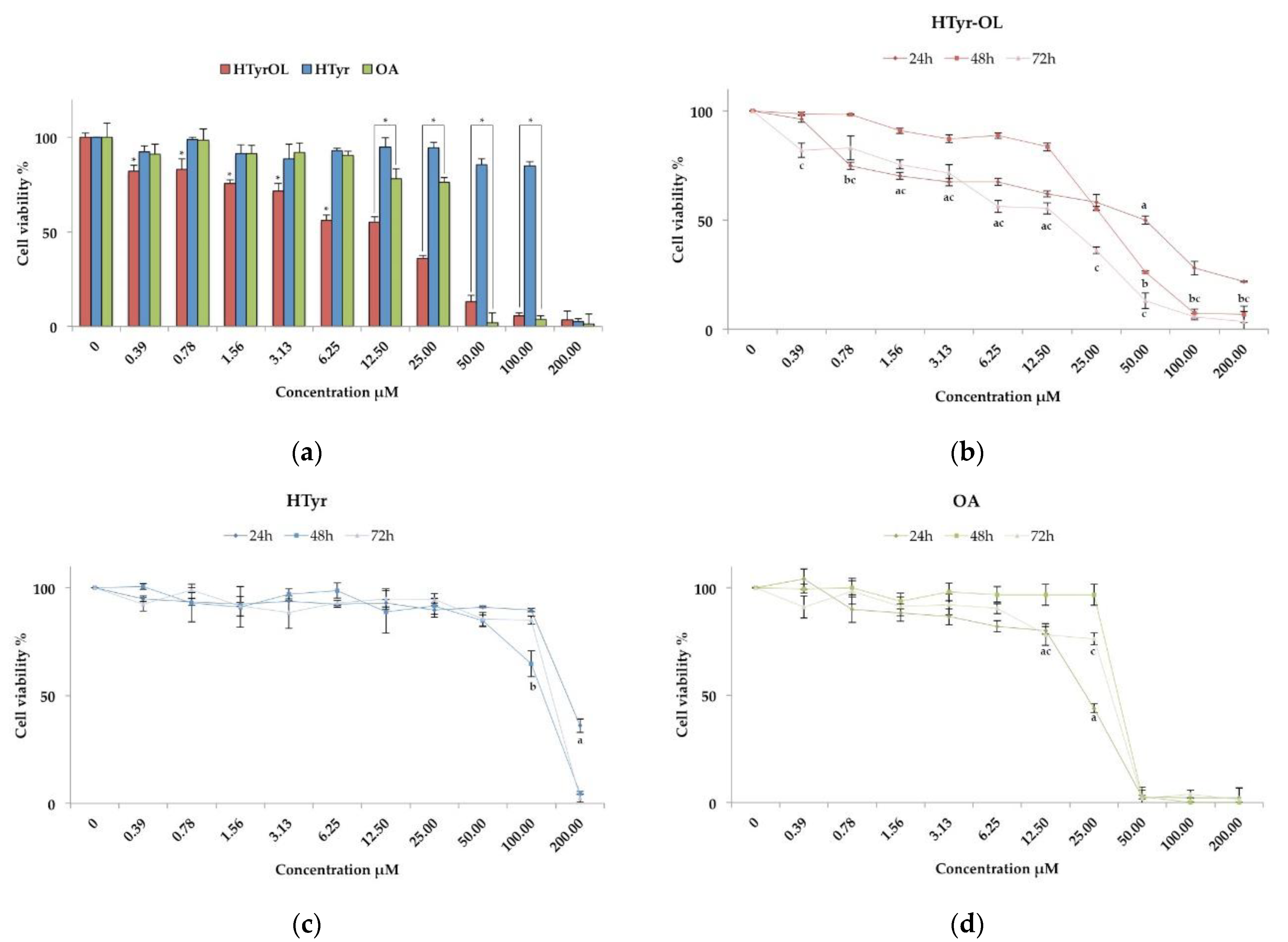
2.2. Cytotoxic Activity
2.3. Anti-Proliferative Effect
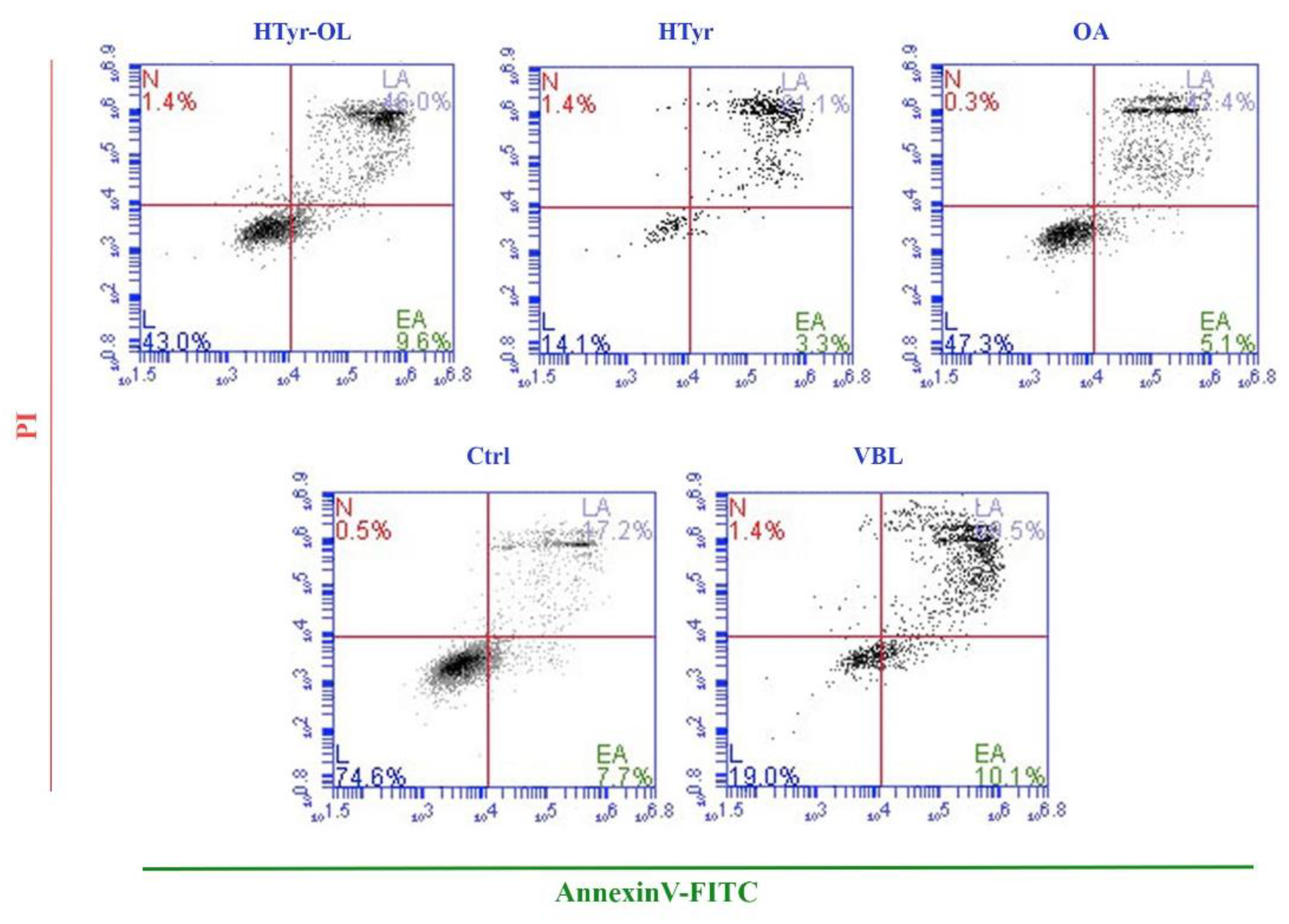
2.4. Apoptosis Detection by Flow Cytometry
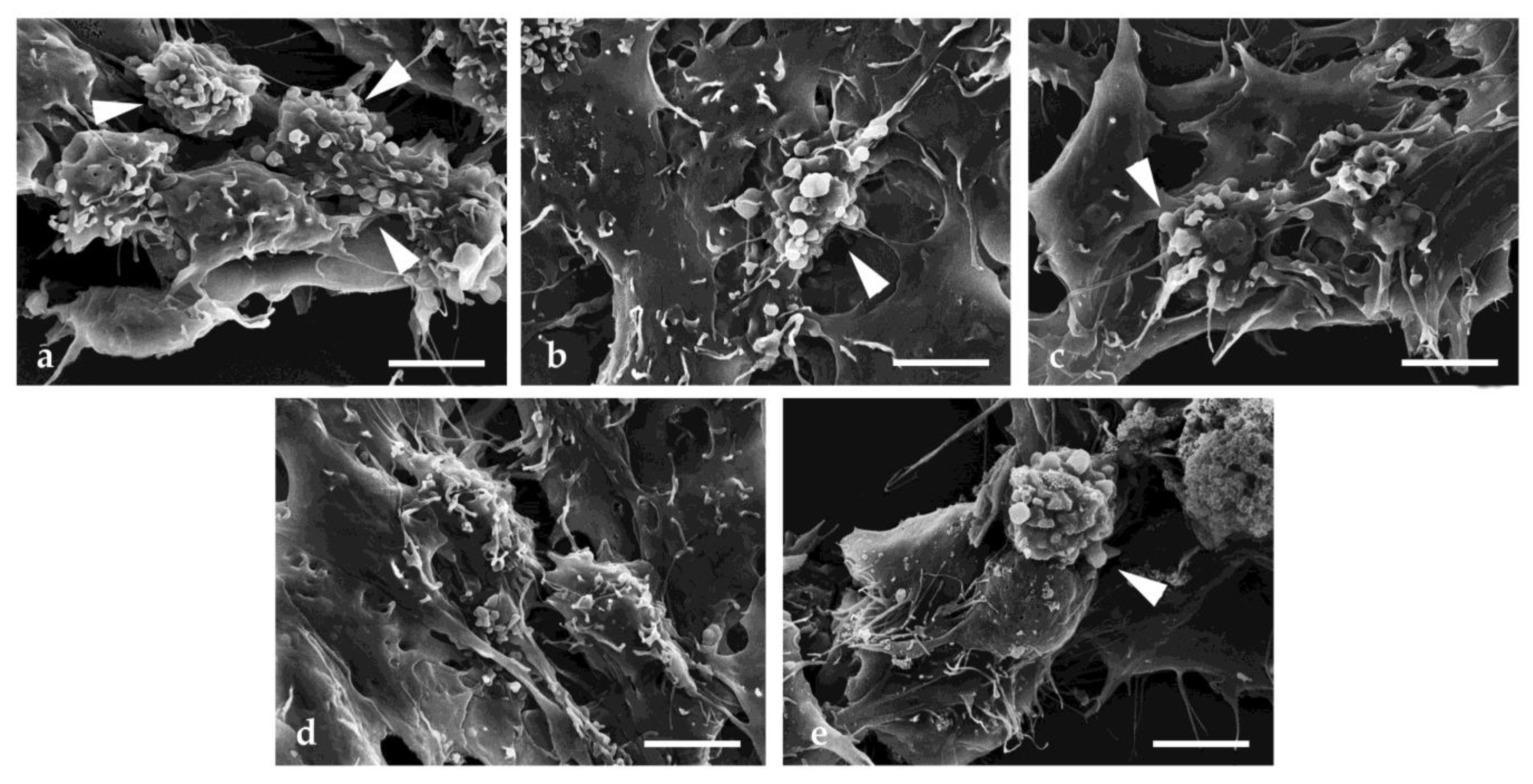
2.5. Scanning Electron Microscopy Investigation
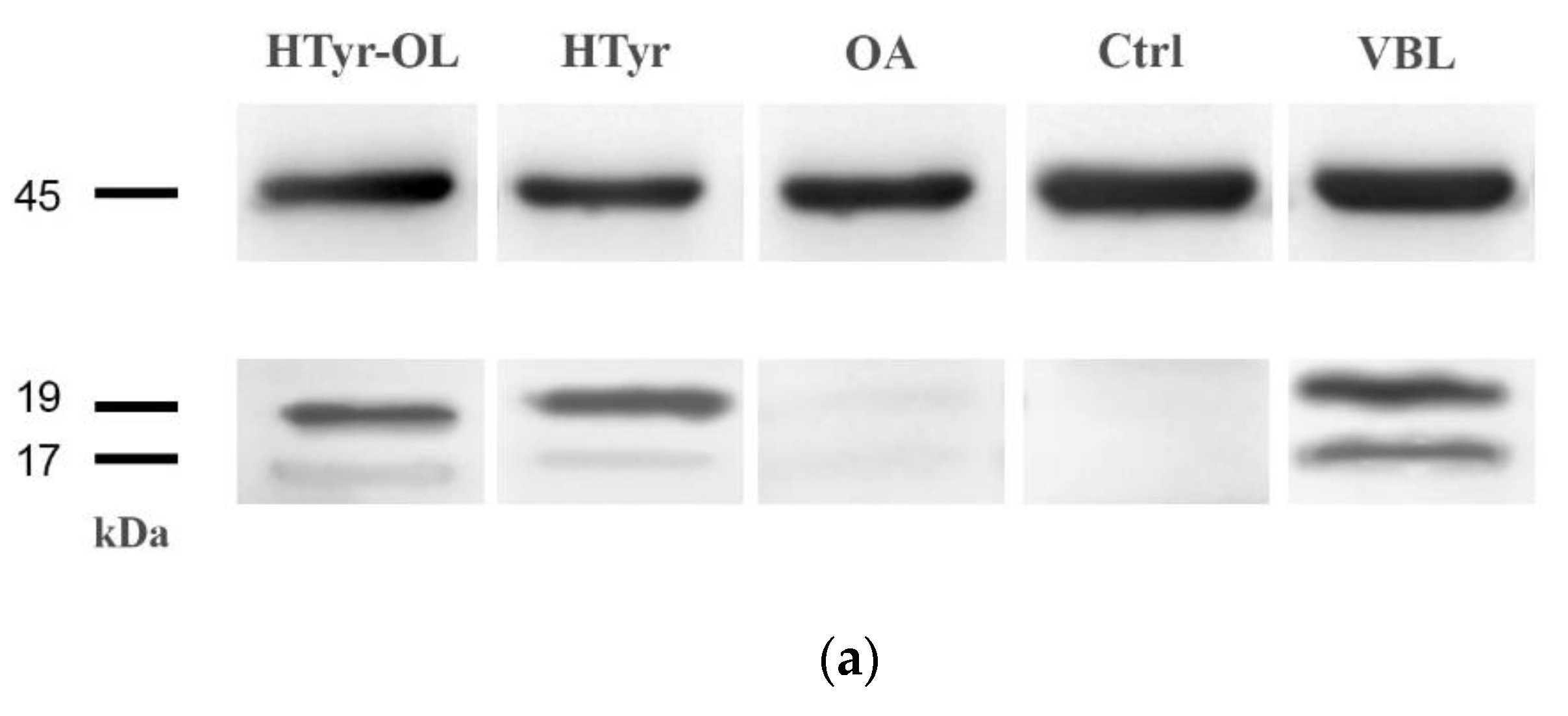
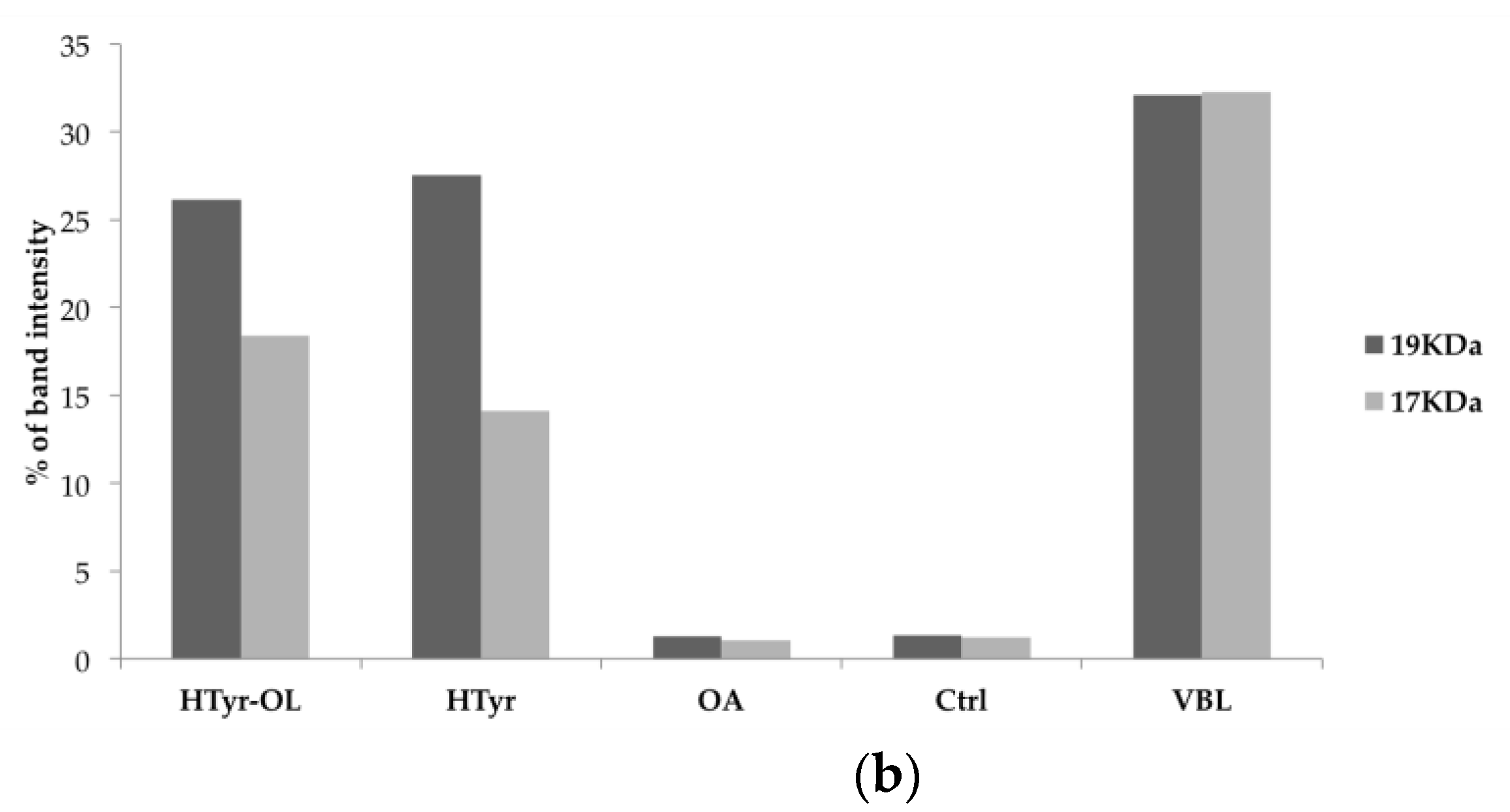
2.6. Detection of Caspase-3 Activated by Western Blot
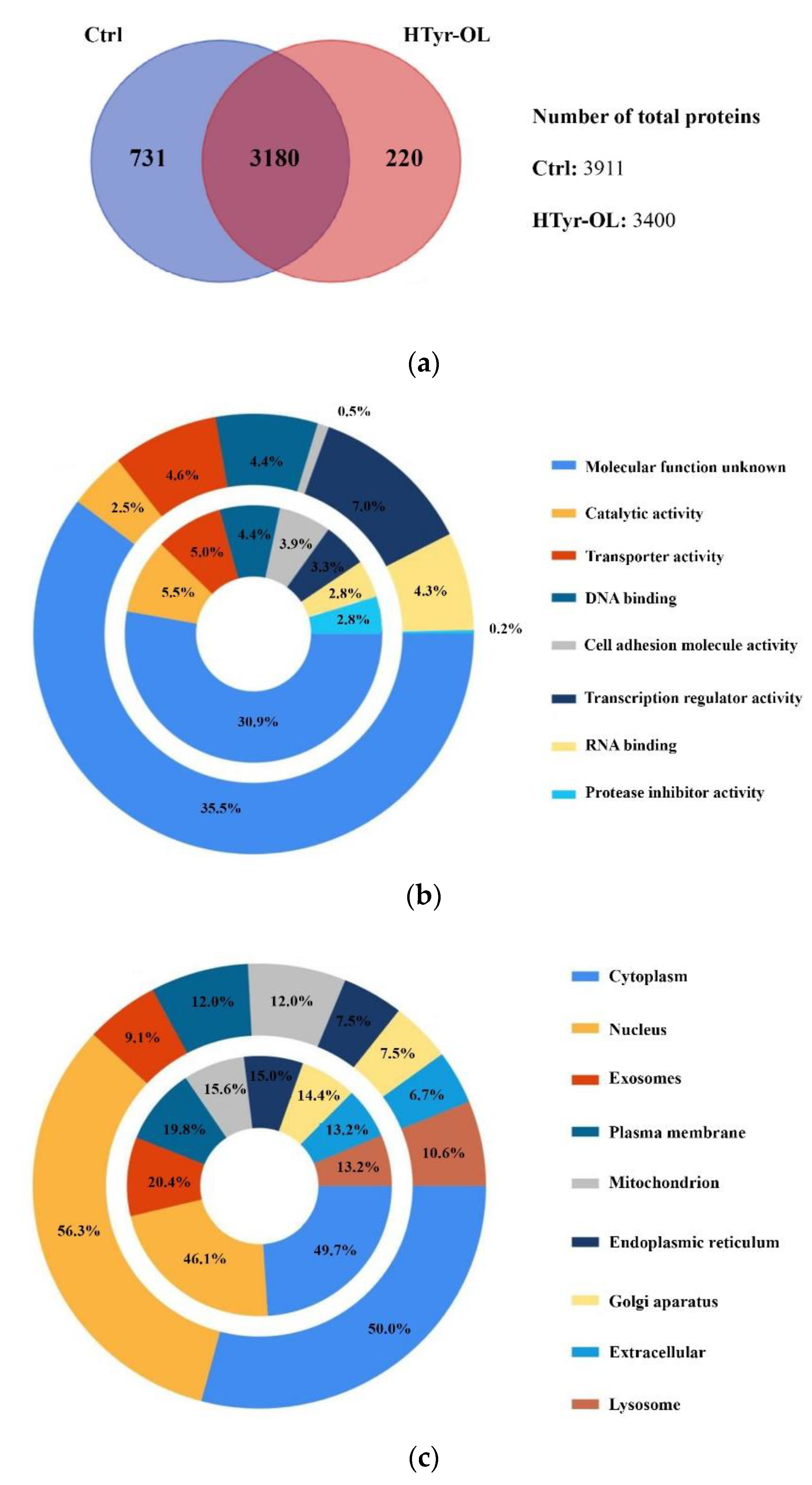
2.7. Shotgun Proteomic Approach and Gene Ontology Functional Annotation
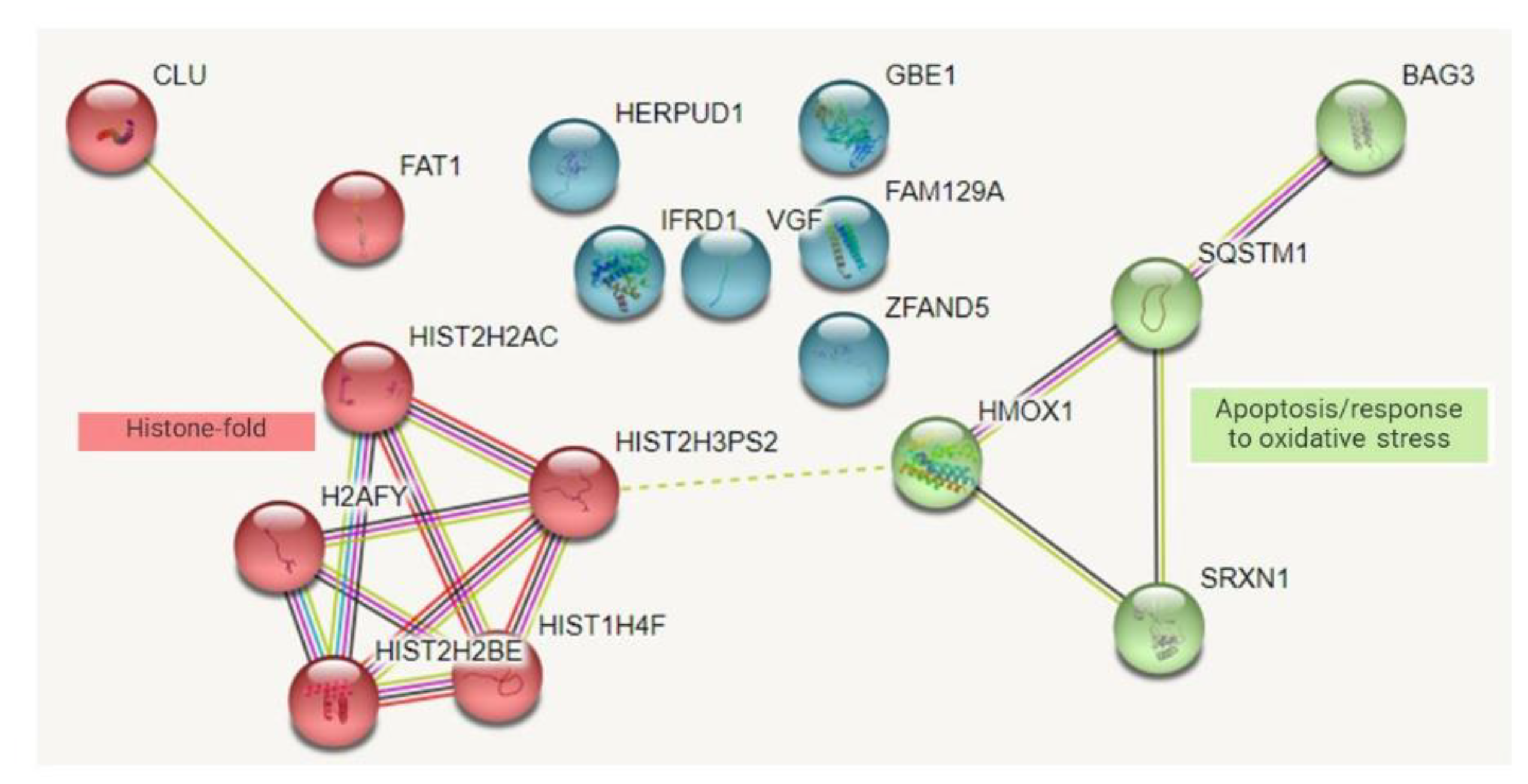
Protein–Protein Interaction Network
3. Discussion
4. Materials and Methods
4.1. Chemical and Reagents
4.2. Instrumentation
4.3. Synthesis of HTyr and HTyr-OL
4.4. Cell Culturing
4.5. Cytotoxicity and Anti-Proliferative Assay
4.6. Apoptosis Detection by Flow Cytometry
4.7. Scanning Electron Microscopy
4.8. Detection of Caspase-3 Activated by Western Blot
4.9. Statystical Analysis
4.10. Proteomic Analysis
4.10.1. Preparation of Cell Culture Samples
4.10.2. Proteomic Analysis
4.10.3. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Bernardini, S.; Tiezzi, A.; Laghezza Masci, V.; Ovidi, E. Natural products for human health: An historical overview of the drug discovery approaches. Nat. Prod. Res. 2018, 32, 1926–1950. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, M.; Kapulnik, Y.; Koltai, H. Plant derived substances with anti-cancer activity: From folklore to practice. Front. Plant Sci. 2015, 6, 799. [Google Scholar] [CrossRef] [PubMed]
- Lichota, A.; Gwozdzinski, K. Anticancer Activity of Natural Compounds from Plant and Marine Environment. Int. J. Mol. Sci. 2018, 19, 3533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najmi, A.; Javed, S.A.; Al Bratty, M.; Alhazmi, H.A. Modern Approaches in the Discovery and Development of Plant-Based Natural Products and Their Analogues as Potential Therapeutic Agents. Molecules 2022, 27, 349. [Google Scholar] [CrossRef] [PubMed]
- Schwingshacki, L.; Morze, J.; Hoffmann, G. Mediterranean diet and health status: Active ingredients and pharmacological mechanisms. Br. J. Pharmacol. 2020, 177, 1241–1257. [Google Scholar] [CrossRef] [Green Version]
- Bonofiglio, D. Mediterranean diet and physical activity as healthy lifestyles for human health. Nutrients 2022, 14, 2514. [Google Scholar] [CrossRef]
- Cory, H.; Passarelli, S.; Szeto, J.; Tamez, M.; Mattei, J. The role of polyphenols in human health and food systems: A mini-review. Front. Nutr. 2018, 5, 87. [Google Scholar] [CrossRef] [Green Version]
- Santos-Buelga, C.; Gonzalez-Manzano, S.; Gonzalez-Paramas, A.M. Wine, polyphenols, and mediterranean diets. What else is there to say? Molecules 2021, 26, 5537. [Google Scholar] [CrossRef]
- Gambacorta, A.; Tofani, D.; Bernini, R.; Migliorini, A. High yielding preparation of a stable precursor of hydroxytyrosol by total synthesis and from the natural glucoside oleuropein. J. Agric. Food Chem. 2007, 55, 3386–3391. [Google Scholar] [CrossRef]
- Faustino, M.; Veiga, M.; Sousa, P.; Costa, E.M.; Silva, S.; Pintado, M. Agro-food byproducts as a new source of natural food sdditives. Molecules 2019, 24, 1056. [Google Scholar] [CrossRef]
- Romani, A.; Pinelli, P.; Ieri, F.; Bernini, R. Sustainability, innovation and green chemistry in the production and valorization of phenolic extracts from Olea europaea L. Sustainability 2016, 8, 1002. [Google Scholar] [CrossRef] [Green Version]
- Luzi, F.; Pannucci, E.; Clemente, M.; Grande, E.; Urciuoli, S.; Romani, A.; Torre, L.; Puglia, D.; Bernini, R.; Santi, L. Hydroxytyrosol and oleuropein-enriched extracts obtained from olive oil wastes and by-products as active antioxidant ingredients for poly-(vinyl alcohol)-based films. Molecules 2021, 26, 2104. [Google Scholar] [CrossRef] [PubMed]
- Aunon-Calles, D.; Giordano, E.; Bohnenberger, S.; Visioli, F. Hydroxytyrosol is not genotoxic in vitro. Pharmacol. Res. 2013, 74, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Aunon-Calles, D.; Canut, L.; Visioli, F. Toxicological evaluation of pure hydroxytyrosol. Food Chem. Toxicol. 2013, 55, 408–504. [Google Scholar] [CrossRef]
- Markovic, A.K.; Toric, J.; Barbaric, M.; Brala, C.J. Hydroxytyrosol, tyrosol and derivatives and their potential effects on human health. Molecules 2019, 24, 2001. [Google Scholar] [CrossRef] [Green Version]
- Fuccelli, R.; Fabiani, R.; Rosignoli, P. Hydroxytyrosol exerts antinflammatory and antioxidant activities in a mouse model of systemic inflammation. Molecules 2018, 23, 3212. [Google Scholar] [CrossRef] [Green Version]
- Leo, M.; Muccillo, L.; Dugo, L.; Bernini, R.; Santi, L.; Sabatino, L. Polyphenols extracts from oil production waste products (OPWPs) reduce cell viability and exert anti-inflammatory activity via PPAγ induction in colorectal cancer cells. Antioxidants 2022, 11, 624. [Google Scholar] [CrossRef]
- Fabiani, R.; Rosignoli, P.; De Bartolomeo, A.; Fuccelli, R.; Morozzi, G. Inhibition of cell cycle progression by hydroxytyrosol is associated with upregulation of cyclindependent protein kinase inhibitors p21(WAF1/Cip1) and p27(Kip1) and with induction of differentiation in HL60 cells. J. Nutr. 2008, 138, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Bernini, R.; Merendino, N.; Romani, A.; Velotti, F. Naturally occurring hydroxytyrosol: Synthesis and anticancer potential. Curr. Med. Chem. 2013, 20, 655–670. [Google Scholar] [CrossRef]
- Bernini, R.; Gilardini Montani, M.S.; Merendino, N.; Romani, A.; Velotti, F. Hydroxytyrosol-derived compounds: A basis for the creation of new pharmacological agents for cancer prevention and therapy. J. Med. Chem. 2015, 58, 9089–9107. [Google Scholar] [CrossRef]
- Hamden, K.; Allouche, N.; Jouadi, B.; El-Fazaa, S.; Gharbi, N.; Carreau, S.; Damak, M.; Elfeki, A. Inhibitory action of purified hydroxytyrosol from stored olive mill waste on intestinal disaccharidases and lipase activities and pancreatic toxicity in diabetic rats. Food Sci. Biotechnol. 2010, 19, 439–447. [Google Scholar] [CrossRef]
- Covas, M.I.; Nyyssonen, K.; Poulsen, H.E.; Kaikkonen, J.; Zunft, H.J.; Kiesewetter, H.; Gaddi, A.; de la Torre, R.; Mursu, J.; Baumler, H.; et al. The effect of polyphenols in olive oil on heart disease risk factors: A randomized trial. Ann. Intern. Med. 2006, 145, 333–341. [Google Scholar] [CrossRef]
- Gonzalez-Santiago, M.; Martin-Bautista, E.; Carrero, J.J.; Fonolla, J.; Baro, L.; Bartolomé, M.V.; Gil-Loyzaga, P.; López-Huertas, E. One-month administration of hydroxytyrosol a phenolic antioxidant present in olive oil to hyperlipemic rabbits improves blood lipid profile antioxidant status and reduces atherosclerosis development. Atherosclerosis 2006, 188, 35–42. [Google Scholar] [CrossRef]
- González-Correa, J.A.; Navas, M.D.; Lopez-Villodres, J.A.; Trujillo, M.; Espartero, J.L.; de La Cruz, J.P. Neuroprotective effect of hydroxytyrosol and hydroxytyrosol acetate in rat brain slices subjected to hypoxia-reoxygenation. Neurosci. Lett. 2008, 446, 143–146. [Google Scholar] [CrossRef]
- Rodriguez-Morato, J.; Xicota, L.; Fito, M.; Farré, M.; Dierssen, M.; de La Torre, R. Potential role of olive oil phenolic compounds in the prevention of neurodegenerative diseases. Molecules 2015, 20, 4655–4680. [Google Scholar] [CrossRef] [Green Version]
- D’Andrea, G.; Ceccarelli, M.; Bernini, R.; Clemente, M.; Santi, L.; Caruso, C.; Micheli, L.; Tirone, F. Hydroxytyrosol stimulates neurogenesis in aged dentate gyrus by enhancing stem and progenitor cell proliferation and neuron survival. FASEB J. 2020, 34, 4512–4526. [Google Scholar] [CrossRef] [Green Version]
- EFSA. Scientific Opinion on the Substantiation of Health Claims Related to Polyphenols in Olive and Protection of LDL Particles from Oxidative Damage (ID 1333,1638, 1639, 1696, 2865), Maintenance of Normal Blood HDL Cholesterol Concentrations (ID 1639), Maintenance of Normal Blood Pressure (ID 3781), “Anti-Inflammatory Properties” (ID 1882), “Contributes to the Upper Respiratory Tract Health” (ID 3468), “Can Help to Maintain a Normal Function of Gastrointestinal Tract” (3779), and “Contributes to Body Defences against External Agents” (ID 3467) Pursuant to Article 13 of Regulation (EC) No 1924/2006. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/2033 (accessed on 5 August 2020).
- Robles-Almazan, M.; Pulido-Moran, M.; Moreno-Fernandez, J.; Ranirez-Tortosa, C.; Rodriguez-Garcia, C.; Quiles, J.L.; Ramirez-Tortosa, M.C. Hydroxytyrosol: Bioavailability, toxicity, and clinical applications. Food Res. Int. 2018, 105, 654–667. [Google Scholar] [CrossRef] [PubMed]
- Bouallagui, Z.; Bouaziz, M.; Lassoued, S.; Engasser, J.M.; Ghoul, M.; Sayadi, S. Hydroxytyrosol acyl esters: Biosynthesis and activities. Appl. Biochem. Biotech. 2011, 163, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M.; Gallardo, E.; Madrona, A.; Bravo, L.; Sarria, B.; Gonzalez-Correa, J.A.; Espartero, J.L. Synthesis and antioxidant activity of nitrohydroxytyrosol and its acyl derivatives. J. Agric. Food Chem. 2014, 62, 10297–10303. [Google Scholar] [CrossRef] [Green Version]
- Bernini, R.; Barontini, M.; Cis, V.; Carastro, I.; Tofani, D.; Chiodo, R.A.; Lupattelli, P.; Incerpi, S. Synthesis and evaluation of the antioxidant activity of lipophilic phenethyl trifluoroacetate esters by in vitro ABTS, DPPH and in cell-culture DCF assays. Molecules 2018, 23, 208. [Google Scholar] [CrossRef] [PubMed]
- Bernini, R.; Carastro, I.; Santoni, F.; Clemente, M. Synthesis of lipophilic esters of tyrosol, homovanillyl alcohol and hydroxytyrosol. Antioxidants 2019, 8, 174. [Google Scholar] [CrossRef] [Green Version]
- Pereira-Caro, G.; Madrona, A.; Bravo, L.; Cert, A.; Espartero, J.L.; Alcudia, F.; Mateos, R. Antioxidant activity evaluation of alkyl hydroxytyrosyl ethers, a new class of hydroxytyrosol derivatives. Food Chem. 2009, 115, 86–91. [Google Scholar] [CrossRef]
- Procopio, A.; Celia, C.; Nardi, M.; Oliverio, M.; Paolino, D.; Sindona, G. Lipophilic hydroxytyrosol esters: Fatty acid conjugates for potential topical administration. J. Nat. Prod. 2011, 74, 2377–2381. [Google Scholar] [CrossRef]
- Calderon-Montano, J.M.; Madrona, A.; Burgos-Moron, E.; Orta, M.L.; Mateos, S.; Espartero, J.L.; Lopez-Lazaro, M. Selective cytotoxic activity of new lipophilic hydroxytyrosol alkyl ether derivatives. J. Agric. Food Chem. 2013, 61, 5046–5053. [Google Scholar] [CrossRef]
- Bianco, A.; Coccioli, F.; Guiso, M.; Marra, C. The occurrence in olive oil of a new class of phenolic compounds: Hydroxy-isochromans. Food Chem. 2002, 77, 405–411. [Google Scholar] [CrossRef]
- Bernini, R.; Crisante, F.; Fabrizi, G.; Gentili, P. Convenient synthesis of 1-aryl-dihydroxyisochromans exhibiting antioxidant activity. Curr. Org. Chem. 2012, 16, 1051–1057. [Google Scholar] [CrossRef]
- Roma, E.; Mattoni, E.; Lupattelli, P.; Moeini, S.S.; Gasperi, T.; Bernini, R.; Incerpi, S.; Tofani, D. New dihydroxytyrosyl esters from dicarboxylic acids. Synthesis and evaluation of the antioxidant activity in vitro (ABTS) and in cell-cultures (DCF assay). Molecules 2020, 25, 3135. [Google Scholar] [CrossRef] [PubMed]
- Bovicelli, P.; Bernini, R.; Antonioletti, R.; Mincione, E. Selective halogenation of flavanones. Tetrahedron Lett. 2002, 43, 5563–5567. [Google Scholar] [CrossRef]
- Plastina, P.; Benincasa, C.; Perri, E.; Fazio, A.; Augimeri, G.; Poland, M.; Witkamp, R.; Meijerink, J. Identification of hydroxytyrosyl oleate, a derivative of hydroxytyrosol with anti-inflammatory properties, in olive oil by-products. Food Chem. 2019, 279, 105–113. [Google Scholar] [CrossRef]
- Bernini, R.; Carastro, I.; Palmini, G.; Tanini, A.; Zonefrati, R.; Pinelli, P.; Brandi, M.L.; Romani, A. Lipophilization of hydroxytyrosol-enriched fractions from Olea europaea L. byproducts and evaluation of the in vitro effects on a model of colorectal cancer cells. J. Agric. Food Chem. 2017, 65, 6506–6512. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, C.; La Torre, C.; Plastina, P.; Fazio, A.; Perri, E.; Caroleo, M.C.; Gallelli, R.; Cannataro, R.; Cione, E. Hydroxytyrosyl oleate: Improved extraction procedure from olive oil and by-products, and in vitro antioxidant and skin regenerative properties. Antioxidants 2019, 8, 233. [Google Scholar] [CrossRef] [Green Version]
- Caroleo, M.C.; Plastina, P.; Fazio, A.; La Torre, C.; Manetti, F. Olive oil lipophenols induce insulin secretion in 832/13 b-cell models. Pharmaceuticals 2021, 13, 1085. [Google Scholar] [CrossRef]
- Gupta, A.; Dwivedi, T. A simplified overview of World Health. Organization classification update of central nervous system tumors 2016. J. Neurosci. Rural Pract. 2017, 8, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernini, R.; Mincione, E.; Barontini, M.; Crisante, F. Convenient synthesis of hydroxytyrosol and its lipophilic derivatives from tyrosol or homovanillyl alcohol. J. Agric. Food Chem. 2008, 56, 8897–8904. [Google Scholar] [CrossRef]
- Bernini, R.; Fabrizi, G.; Pouysegu, L.; Deffieux, D.; Quideau, S. Synthesis of biologically active catecholic compounds via ortho-selective oxygenation of phenolic compounds using hypervalent iodine(V) reagents. Curr. Org. Synth. 2012, 9, 650–669. [Google Scholar] [CrossRef]
- The UniProt Consortium, UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, 480–489. [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A Review of Programmed Cell Death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Saraste, A.; Pulkki, K. Morphologic and biochemical hallmarks of apoptosis. Cardiovasc. Res. 2000, 45, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Bassani, B.; Rossi, T.; De Stefano, D.; Pizzichini, D.; Corradino, P.; Macrì, N.; Noonan, D.M.; Albini, A.; Bruno, A. Potential chemopreventive activities of a polyphenol rich purified extract from olive mill wastewater on colon cancer cells. J. Funct. Foods 2016, 27, 236–248. [Google Scholar] [CrossRef]
- Han, J.; Talorete, T.P.N.; Yamada, P.; Isoda, H. Anti-proliferative and apoptotic effects of oleuropein and hydroxytyrosol on human breast cancer MCF-7 cells. Cytotechnology 2009, 59, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Ragione, F.D.; Cucciolla, V.; Borriello, A.; Pietra, V.D.; Pontoni, G.; Racioppi, L.; Manna, C.; Galletti, P.; Zappia, V. Hydroxytyrosol, a natural molecule occurring in olive oil, induces cytochrome c-dependent apoptosis. Biochem. Biophys. Res. Commun. 2000, 278, 733–739. [Google Scholar] [CrossRef]
- Goldsmith, C.D.; Bond, D.R.; Jankowski, H.; Weidenhofer, J.; Stathopoulos, C.E.; Roach, P.D.; Scarlett, C.J. The Olive Biophenols Oleuropein and Hydroxytyrosol Selectively Reduce Proliferation, Influence the Cell Cycle, and Induce Apoptosis in Pancreatic Cancer Cells. Int. J. Mol. Sci. 2018, 19, 1937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corona, G.; Deiana, M.; Incani, A.; Vauzour, D.; Dessì, M.A.; Spencer, J.P. Hydroxytyrosol inhibits the proliferation of human colon adenocarcinoma cells through inhibition of ERK1/2 and cyclin D1. Mol. Nutr. Food Res. 2009, 53, 897–903. [Google Scholar] [CrossRef]
- Terzuoli, E.; Donnini, S.; Giachetti, A.; Iñiguez, M.A.; Fresno, M.; Melillo, G.; Ziche, M. Inhibition of hypoxia inducible factor-1alpha by dihydroxyphenylethanol, a product from olive oil, blocks microsomal prostaglandin-E synthase-1/vascular endothelial growth factor expression and reduces tumor angiogenesis. Clin. Cancer Res. 2010, 16, 4207–4216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Luo, C.; Liu, J. Hydroxytyrosol induces apoptosis in human colon cancer cells through ROS generation. Food Funct. 2014, 5, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Achmon, Y.; Fishman, A. The antioxidant hydroxytyrosol: Biotechnological production challenges and opportunities. Appl. Microbiol. Biotechnol. 2015, 99, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Schwarz, S.; Ahlemeyer, B.; Grzeschik, S.; Klumpp, S.; Krieglstein, J. OA causes apoptosis and dephosphorylates Bad. Neurochem. Int. 2005, 46, 127–135. [Google Scholar] [CrossRef]
- Leist, M.; Jäättelä, M. Four deaths and a funeral: From caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2001, 2, 589–598. [Google Scholar] [CrossRef]
- Carrillo, C.; Cavia, M.M.; Alonso-Torre, S.R. Antitumor effect of oleic acid; mechanisms of action. A review. Nutr. Hosp. 2012, 27, 1860–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.Q.; Rajadurai, P.; Abas, F.; Othman, I.; Naidu, R. Proteomic Analysis on Anti-Proliferative and Apoptosis Effects of Curcumin Analog, 1,5-bis(4-Hydroxy-3-Methyoxyphenyl)-1,4-Pentadiene-3-One-Treated Human Glioblastoma and Neuroblastoma Cells. Front. Mol. Biosci. 2021, 8, 645856. [Google Scholar] [CrossRef]
- Maurer, U.; Charvet, C.; Wagman, A.S.; Dejardin, E.; Green, D.R. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol. Cell 2006, 21, 749–760. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Xu, Y.; Xia, Q.J.; Zhao, X.M.; Li, S.; He, X.Q.; Wang, R.R.; Wang, T.H. Combined Scutellarin and C18H17NO6 Imperils the Survival of Glioma: Partly Associated With the Repression of PSEN1/PI3K-AKT Signaling Axis. Front. Oncol. 2021, 11, 663262. [Google Scholar] [CrossRef]
- Baki, L.; Shioi, J.; Wen, P.; Shao, Z.; Schwarzman, A.; Gama-Sosa, M.; Neve, R.; Robakis, N.K. PS1 activates PI3K thus inhibiting GSK-3 activity and tau overphosphorylation: Effects of FAD mutations. EMBO J. 2004, 23, 2586–2596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, L.; Zeng, Y.; Wang, J.; Liu, Z.; Shen, B.; Ge, J.; Liu, Y.; Guo, Y.; Qiu, J. Differential microRNA expression in aristolochic acid-induced upper urothelial tract cancers ex vivo. Mol. Med. Rep. 2015, 12, 6533–6546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, V.; Pauleau, A.L.; Steffen, N.; Bergner, A.; Becker, P.B.; Erhardt, S. The histone-fold protein CHRAC14 influences chromatin composition in response to DNA damage. Cell Rep. 2014, 7, 321–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frigerio, M.; Santagostino, M.; Sputore, S. A user-friendly entry to 2-iodoxybenzoic acid (IBX). J. Org. Chem. 1999, 64, 4537–4538. [Google Scholar] [CrossRef]
- Ovidi, E.; Laghezza Masci, V.; Garzoli, S.; Gambellini, G.; Keita, S.V.; Zago, D.; Turchetti, G.; Modesti, L.; Tiezzi, A. Antiproliferative properties of Papaver rhoeas ovule extracts and derived fractions tested on HL60 Leukemia human cells. Molecules 2020, 25, 1850. [Google Scholar] [CrossRef] [Green Version]
- Karnovsky, M.J. A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 1965, 27, 137–138. [Google Scholar]
- Laghezza Masci, V.; Ovidi, E.; Taddei, A.R.; Turchetti, G.; Tiezzi, A.; Giacomello, P.; Garzoli, S. Apoptotic effects on HL60 human leukaemia cells induced by lavandin essential oil treatment. Molecules 2020, 25, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Rhee, S.Y.; Wood, V.; Dolinski, K.; Draghici, S. Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 2008, 9, 509–515. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Chisanga, D.; Alessandro, R.; Ang, C.S.; Askenase, P.; Batagov, A.O.; Benito-Martin, A.; Camussi, G.; Clayton, A.; et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles 2017, 6, 1321455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]

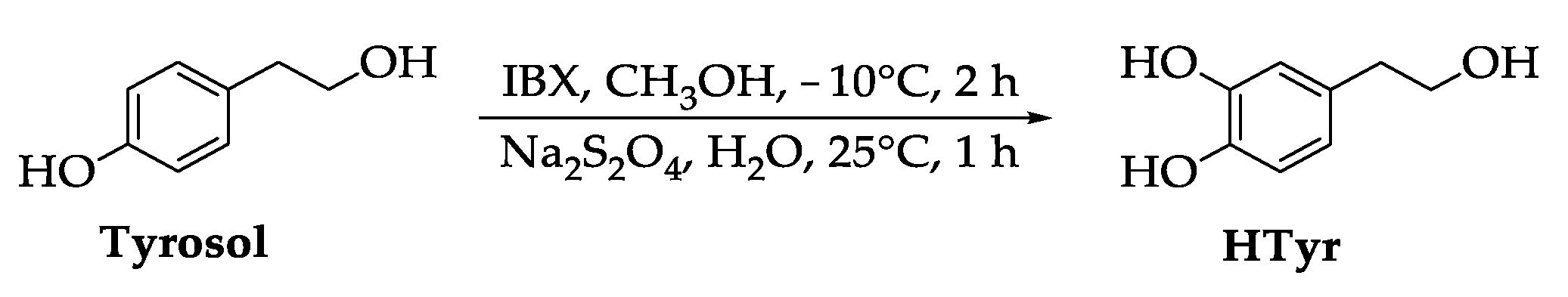

| EC50 (µM Mean ± SD) | ||
|---|---|---|
| Compound | SH-SY5Y | MCF10A |
| HTyr-OL | 26.35 ± 3.75 a | >200 |
| HTyr | 114.02 ± 1.69 | >200 |
| OA | 35.93 ± 3.57 b | >200 |
| EC50 (µM Mean ± SD) | |||
|---|---|---|---|
| Compound | 24 h | 48 h | 72 h |
| HTyr-OL | 48.97 ± 9.04 a | 27.76 ± 2.31 b | 26.35 ± 3.75 b |
| HTyr | 150.13 ± 7.89 | 113.59 ± 7.87 | 114.02 ± 1.69 |
| OA | 36.91 ± 4.93 a | 38.64 ± 5.28 c | 35.93 ± 3.57 c |
| Compound | Apoptotic Rate % |
|---|---|
| HTyr-OL | 52.36 ± 4.31 |
| HTyr | 79.64 ± 8.83 |
| OA | 49.91 ± 6.94 |
| Ctrl | 22.25 ± 2.69 |
| VBL | 74.24 ± 6.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laghezza Masci, V.; Bernini, R.; Villanova, N.; Clemente, M.; Cicaloni, V.; Tinti, L.; Salvini, L.; Taddei, A.R.; Tiezzi, A.; Ovidi, E. In Vitro Anti-Proliferative and Apoptotic Effects of Hydroxytyrosyl Oleate on SH-SY5Y Human Neuroblastoma Cells. Int. J. Mol. Sci. 2022, 23, 12348. https://doi.org/10.3390/ijms232012348
Laghezza Masci V, Bernini R, Villanova N, Clemente M, Cicaloni V, Tinti L, Salvini L, Taddei AR, Tiezzi A, Ovidi E. In Vitro Anti-Proliferative and Apoptotic Effects of Hydroxytyrosyl Oleate on SH-SY5Y Human Neuroblastoma Cells. International Journal of Molecular Sciences. 2022; 23(20):12348. https://doi.org/10.3390/ijms232012348
Chicago/Turabian StyleLaghezza Masci, Valentina, Roberta Bernini, Noemi Villanova, Mariangela Clemente, Vittoria Cicaloni, Laura Tinti, Laura Salvini, Anna Rita Taddei, Antonio Tiezzi, and Elisa Ovidi. 2022. "In Vitro Anti-Proliferative and Apoptotic Effects of Hydroxytyrosyl Oleate on SH-SY5Y Human Neuroblastoma Cells" International Journal of Molecular Sciences 23, no. 20: 12348. https://doi.org/10.3390/ijms232012348






