Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant
Abstract
:1. Introduction
2. Results
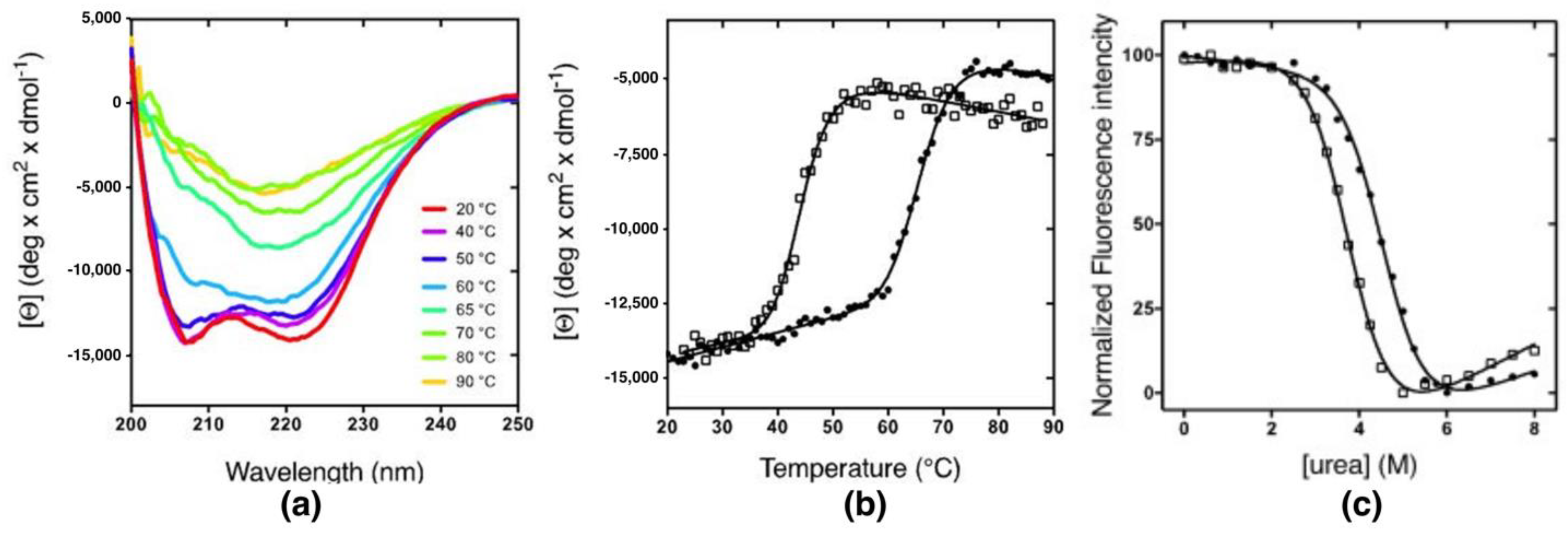
2.1. Equilibrium Experiments
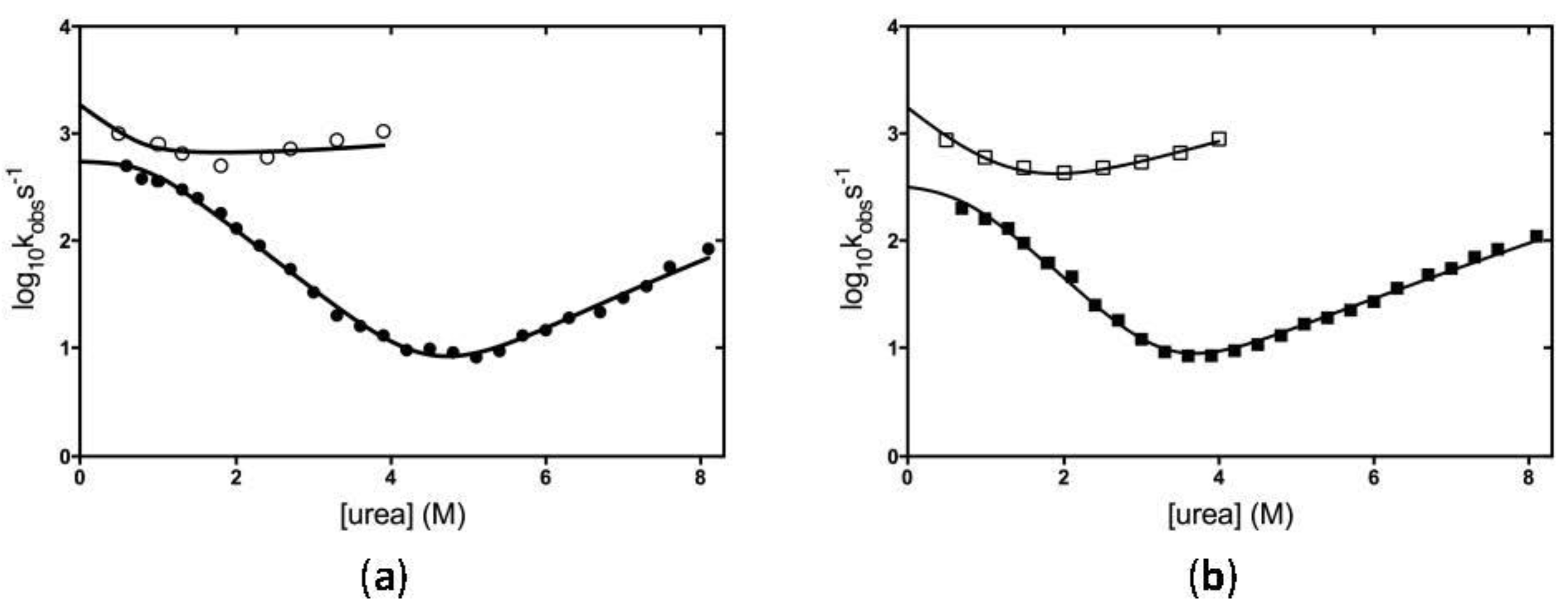
2.2. The Folding Mechanism of KH0 and KH0-R138Q
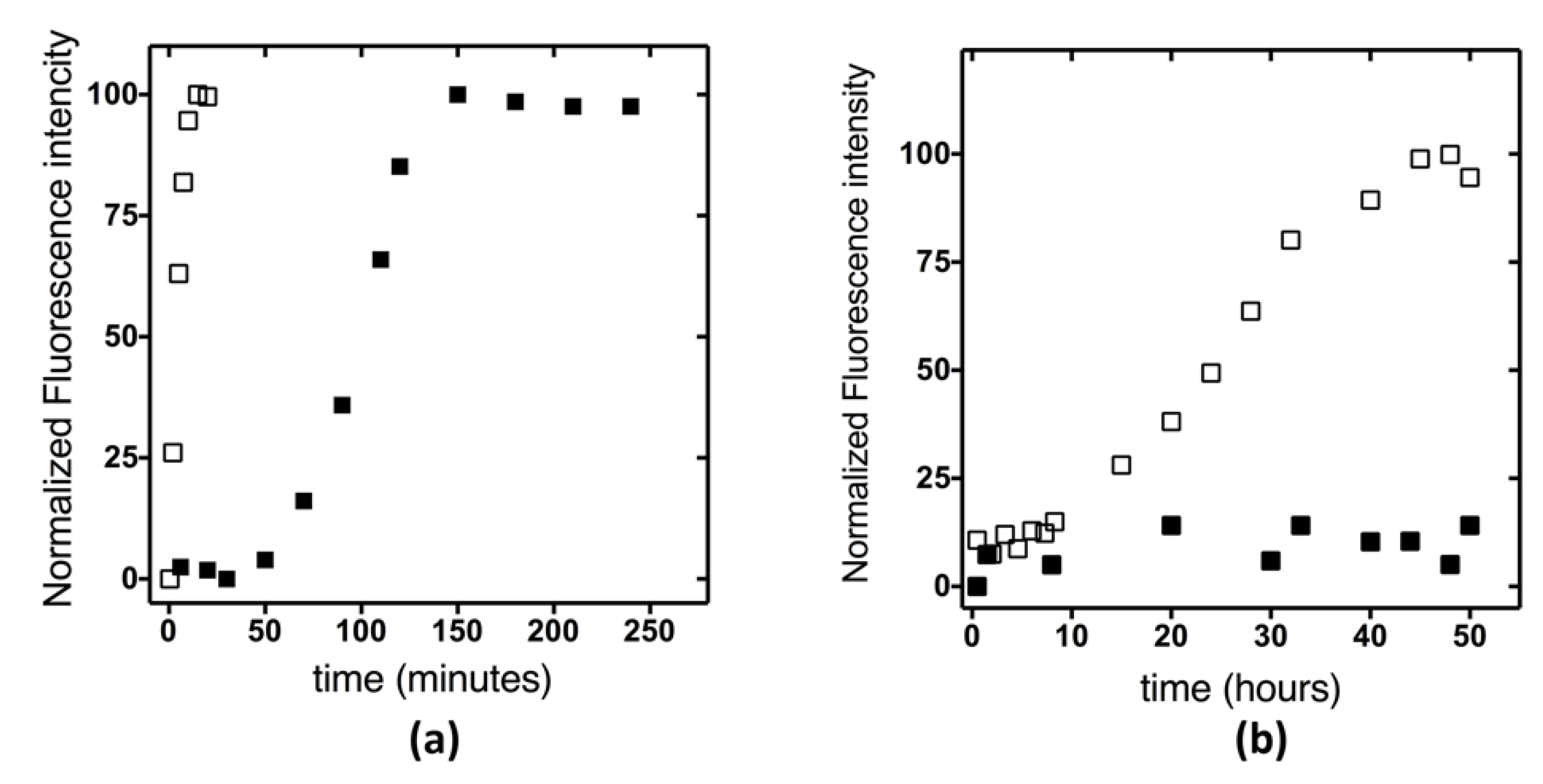
2.3. Aggregation and Fibrillogenesis Propensity of KH0 and KH0-R138Q
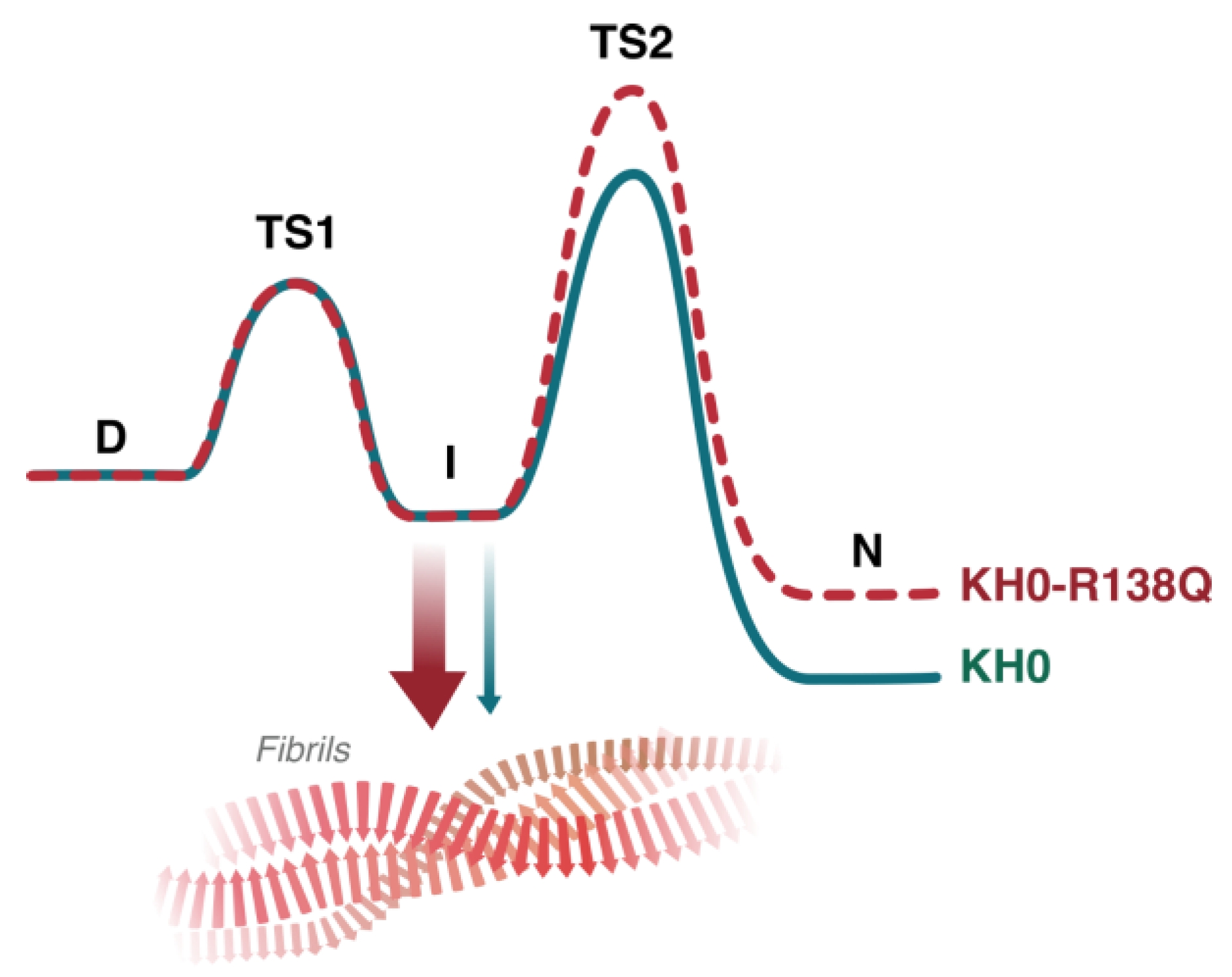
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Equilibrium Experiments
4.3. Kinetic (Un)Folding Experiments
4.4. Thioflavin T Fluorescence Assay
4.5. TEM Experiments
4.6. X-ray Diffraction
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siomi, H.; Matunis, M.J.; Michael, W.M.; Dreyfuss, G. The pre-mRNA binding K protein contains a novel evolutionary conserved motif. Nucleic Acids Res. 1993, 21, 1193–1198. [Google Scholar] [CrossRef]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef]
- Nicastro, G.; Taylor, I.A.; Ramos, A. KH-RNA interactions: Back in the groove. Curr. Opin. Struct. Biol. 2015, 30, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Hollingworth, D.; Candel, A.M.; Nicastro, G.; Martin, S.R.; Briata, P.; Gherzi, R.; Ramos, A. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012, 40, 6873–6886. [Google Scholar] [CrossRef]
- Pereira, J.; Lupas, A.N. The ancestral KH peptide at the root of a domain family with three different folds. Bioinformatics 2018, 34, 3961–3965. [Google Scholar] [CrossRef]
- Santorelli, D.; Rocchio, S.; Fata, F.; Silvestri, I.; Angelucci, F.; Imperi, F.; Marasco, D.; Diaferia, C.; Gigli, L.; Demitri, N.; et al. The folding and aggregation properties of a single KH-domain protein: Ribosome binding factor A (RbfA) from Pseudomonas aeruginosa. Biochim. Biophys. Acta—Gen. Subj. 2021, 1865, 129780. [Google Scholar] [CrossRef]
- Silva, A.P.G.; Chechik, M.; Byrne, R.T.; Waterman, D.G.; Ng, C.L.; Dodson, E.J.; Koonin, E.V.; Antson, A.A.; Smits, C. Structure and activity of a novel archaeal β-CASP protein with N-terminal KH domains. Structure 2011, 19, 622–632. [Google Scholar] [CrossRef] [Green Version]
- Chmiel, N.H.; Rio, D.C.; Doudna, J.A. Distinct contributions of KH domains to substrate binding affinity of Drosophila P-element somatic inhibitor protein. RNA 2006, 12, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.H.K.; Jansen, R.P. A jack of all trades: The RNA-binding protein vigilin. RNA 2017, 8, e1448. [Google Scholar] [CrossRef]
- Oddone, A.; Lorentzen, E.; Basquin, J.; Gasch, A.; Rybin, V.; Conti, E.; Sattler, M. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007, 8, 63–69. [Google Scholar] [CrossRef]
- Myrick, L.K.; Hashimoto, H.; Cheng, X.; Warren, S.T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 2015, 24, 1733–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Annessa, I.; Cicconardi, F.; di Marino, D. Handling FMRP and its molecular partners: Structural insights into Fragile X Syndrome. Prog. Biophys. Mol. Biol. 2019, 141, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.K.; Broadie, K. Multifarious Functions of the Fragile X Mental Retardation Protein. Trends Genet. 2017, 33, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Starke, E.L.; Zius, K.; Barbee, S.A. FXS causing missense mutations disrupt FMRP granule formation, dynamics, and function. PloS Genet. 2022, 18, e1010084. [Google Scholar] [CrossRef]
- Lai, A.; Valdez-Sinon, A.N.; Bassell, G.J.; Gary Bassell, C.J. Regulation of RNA granules by FMRP and implications for neurological diseases. Traffic 2020, 2, 454–462. [Google Scholar] [CrossRef]
- Gareau, C.; Martel, D.; Coudert, L.; Mellaoui, S.; Mazroui, R. Characterization of fragile X mental retardation protein granules formation and dynamics in Drosophila. Biol. Open 2013, 2, 68–81. [Google Scholar] [CrossRef] [Green Version]
- Adinolfi, S.; Bagni, C.; Musco, G.; Gibson, T.; Mazzarella, L.; Pastore, A. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 1999, 5, 1248–1258. [Google Scholar] [CrossRef] [Green Version]
- Musco, G.; Kharrat, A.; Fraternali, F.; Gibson, T.J.; Nilgen, M.; Pastore, A. The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 1997, 4, 712–716. [Google Scholar] [CrossRef]
- Musco, G.; Stier, G.; Joseph, C.; Morelli, M.A.C.; Nilges, M.; Gibson, T.J.; Pastore, A. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 1996, 85, 237–245. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Berry-Kravis, E.; Hazlett, H.C.; Bailey, D.B.; Moine, H.; Kooy, R.F.; Tassone, F.; Gantois, I.; Sonenberg, N.; Mandel, J.L.; et al. Fragile X syndrome. Nat. Rev. Dis. Prim. 2017, 3, 17065. [Google Scholar] [CrossRef] [PubMed]
- Sitzmann, A.F.; Hagelstrom, R.T.; Tassone, F.; Hagerman, R.J.; Butler, M.G. Rare FMR1 gene mutations causing fragile X syndrome: A review. Am. J. Med. Genet. Part A 2018, 176, 11–18. [Google Scholar] [CrossRef]
- Quartier, A.; Poquet, H.; Gilbert-Dussardier, B.; Rossi, M.; Casteleyn, A.S.; Portes, V.d.; Feger, C.; Nourisson, E.; Kuentz, P.; Redin, C.; et al. Intragenic FMR1 disease-causing variants: A significant mutational mechanism leading to Fragile-X syndrome. Eur. J. Hum. Genet. 2017, 25, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, S.C.; Bray, S.M.; Suhl, J.A.; Cutler, D.J.; Coffee, B.; Zwick, M.E.; Warren, S.T. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am. J. Med. Genet. A 2010, 152, 2512–2520. [Google Scholar] [CrossRef] [Green Version]
- Diaz, J.; Scheiner, C. Leon E Presentation of a recurrent FMR1 missense mutation (R138Q) in an affected female. Transl. Sci. Rare Dis. 2018, 3, 139–144. [Google Scholar]
- Myrick, L.K.; Deng, P.Y.; Hashimoto, H.; Oh, Y.M.; Cho, Y.; Poidevin, M.J.; Suhl, J.A.; Visootsak, J.; Cavalli, V.; Jin, P.; et al. Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. Proc. Natl. Acad. Sci. USA 2015, 112, 949–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suhl, J.A.; Warren, S.T. Single-Nucleotide mutations in FMR1 reveal novel functions and regulatory mechanisms of the fragile X syndrome protein FMRP. J. Exp. Neurosci. 2015, 9, 35–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto, M.; Folci, A.; Poupon, G.; Schiavi, S.; Buzzelli, V.; Pronot, M.; François, U.; Pousinha, P.; Lattuada, N.; Abelanet, S.; et al. Missense mutation of Fmr1 results in impaired AMPAR-mediated plasticity and socio-cognitive deficits in mice. Nat. Commun. 2021, 12, 1557. [Google Scholar] [CrossRef] [PubMed]
- Sjekloća, L.; Pauwels, K.; Pastore, A. On the aggregation properties of FMRP—A link with the FXTAS syndrome? FEBS J. 2011, 278, 1912–1921. [Google Scholar] [CrossRef]
- Parker, M.J.; Spencer, J.; Clarke, A.R. An Integrated Kinetic Analysis of Intermediates and Transition States in Protein Folding Reactions. J. Mol. Biol. 1995, 253, 771–786. [Google Scholar] [CrossRef]
- Gianni, S.; Ivarsson, Y.; Jemth, P.; Brunori, M.; Travaglini-Allocatelli, C. Identification and characterization of protein folding intermediates. Biophys. Chem. 2007, 128, 105–113. [Google Scholar] [CrossRef]
- Malgieri, G.; D’Abrosca, G.; Pirone, L.; Toto, A.; Palmieri, M.; Russo, L.; Sciacca, M.F.M.; Tatè, R.; Sivo, V.; Baglivo, I.; et al. Folding mechanisms steer the amyloid fibril formation propensity of highly homologous proteins. Chem. Sci. 2018, 9, 3290–3298. [Google Scholar] [CrossRef] [Green Version]
- del Poggetto, E.; Chiti, F.; Bemporad, F. The Folding process of Human Profilin-1, a novel protein associated with familial amyotrophic lateral sclerosis. Sci. Rep. 2015, 5, 12332. [Google Scholar] [CrossRef] [Green Version]
- Neudecker, P.; Robustelli, P.; Cavalli, A.; Walsh, P.; Lundström, P.; Zarrine-Afsar, A.; Sharpe, S.; Vendruscolo, M.; Kay, L.E. Structure of an Intermediate State in Protein Folding and Aggregation. Science 2012, 336, 362–366. [Google Scholar] [CrossRef]
- Chakraborty, I.; Kar, R.K.; Sarkar, D.; Kumar, S.; Maiti, N.C.; Mandal, A.K.; Bhunia, A. Solvent Relaxation NMR: A Tool for Real-Time Monitoring Water Dynamics in Protein Aggregation Landscape. ACS Chem. Neurosci. 2021, 12, 2903–2916. [Google Scholar] [CrossRef]
- Nilsson, M.R. Techniques to study amyloid fibril formation in vitro. Methods 2004, 34, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Sulatskaya, A.I.; Maskevich, A.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Fluorescence Quantum Yield of Thioflavin T in Rigid Isotropic Solution and Incorporated into the Amyloid Fibrils. PLoS ONE 2010, 5, e15385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldsbury, C.; Baxa, U.; Simon, M.N.; Steven, A.C.; Engel, A.; Wall, J.S.; Aebi, U.; Müller, S.A. Amyloid structure and assembly: Insights from scanning transmission electron microscopy. J. Struct. Biol. 2011, 173, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novak, E.; Petrosino, M.; Santorelli, D.; Chiaraluce, R.; Consalvi, V.; Pasquo, A.; Travaglini-Allocatelli, C. A Glimpse into the Structural Properties of the Intermediate and Transition State in the Folding of Bromodomain 2 Domain 2 by Φ Value Analysis. Int. J. Mol. Sci. 2021, 22, 5953. [Google Scholar] [CrossRef]
- Fersht, A.R.; Sato, S. Value analysis and the nature of protein-folding transition states. Proc. Natl. Acad. Sci. USA 2004, 101, 7976–7981. [Google Scholar] [CrossRef]
- Zheng, W.; Tsai, M.Y.; Chen, M.; Wolynes, P.G. Exploring the aggregation free energy landscape of the amyloid-β protein (1–40). Proc. Natl. Acad. Sci. USA 2016, 113, 11835–11840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Wolynes, P.G. Aggregation landscapes of Huntingtin exon 1 protein fragments and the critical repeat length for the onset of Huntington’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, 4406–4411. [Google Scholar] [CrossRef] [Green Version]
- Grishin, N.V. Fold change in evolution of protein structures. J. Struct. Biol. 2001, 134, 167–185. [Google Scholar] [CrossRef] [Green Version]
- Grishin, N.V. KH domain: One motif, two folds. Nucleic Acids Res. 2001, 29, 638–643. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lausi, A.; Polentarutti, M.; Onesti, S.; Plaisier, J.R.; Busetto, E.; Bais, G.; Barba, L.; Cassetta, A.; Campi, G.; Lamba, D.; et al. Status of the crystallography beamlines at Elettra. Eur. Phys. J. Plus 2015, 130, 43. [Google Scholar] [CrossRef]
- Hammersley, A.P. FIT2D: A multi-purpose data reduction, analysis and visualization program. J. Appl. Crystallogr. 2016, 49, 646–652. [Google Scholar] [CrossRef]
- Huang, T.C.; Toraya, H.; Blanton, T.N.; Wu, Y. X-ray Powder Diffraction Analysis of Silver Behenate, a Possible Low-Angle Diffraction Standard. J. Appl. Cryst. 1993, 26, 180–184. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.; Bolen, D.W. Unfolding Free Energy Changes Determined by the Linear Extrapolation Method. 1. Unfolding Phenylmethanesulfonyl a-Chymotrypsin Using Different Denaturants. Biochemistry 1988, 27, 8063–8068. [Google Scholar] [CrossRef] [PubMed]


| Circular Dichroism | Fluorescence | |||
|---|---|---|---|---|
| KH0 | KH0-R138Q | W-KH0 | W-KH0 R138Q | |
| [Urea]1/2 | 4.30 ± 0.04 | 3.46 ± 0.06 | 4.56 ± 0.04 | 3.77 ± 0.04 |
| m | 1.29 ± 0.06 | 1.29 ± 0.06 | 1.25 ± 0.06 | 1.25 ± 0.06 |
| DGD-N | 5.50 ± 0.20 | 4.46 ± 0.23 | 5.70 ± 0.32 | 4.70 ± 0.24 |
| W-KH0 | W-KH0 R138Q | |
|---|---|---|
| kDI | 1860 ± 220 | 1723 ± 123 |
| mDI | 0.75 ± 0.04 | 0.87 ± 0.02 |
| kID | 29 ± 20 | 54 ± 6 |
| mID | 0.22 ± 0.10 | 0.33 ± 0.03 |
| kIN | 584 ± 58 | 243 ± 12 |
| mIN | 10−12 ± 10−3 | 10−12 ± 10−3 |
| kNI | 1.62 ± 1.1 | 1.20 ± 0.04 |
| mNI | 0.32 ± 0.06 | 0.32 (fixed) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santorelli, D.; Troilo, F.; Fata, F.; Angelucci, F.; Demitri, N.; Giardina, G.; Federici, L.; Catalano, F.; Di Matteo, A.; Travaglini-Allocatelli, C. Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant. Int. J. Mol. Sci. 2022, 23, 12178. https://doi.org/10.3390/ijms232012178
Santorelli D, Troilo F, Fata F, Angelucci F, Demitri N, Giardina G, Federici L, Catalano F, Di Matteo A, Travaglini-Allocatelli C. Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant. International Journal of Molecular Sciences. 2022; 23(20):12178. https://doi.org/10.3390/ijms232012178
Chicago/Turabian StyleSantorelli, Daniele, Francesca Troilo, Francesca Fata, Francesco Angelucci, Nicola Demitri, Giorgio Giardina, Luca Federici, Flavia Catalano, Adele Di Matteo, and Carlo Travaglini-Allocatelli. 2022. "Folding Mechanism and Aggregation Propensity of the KH0 Domain of FMRP and Its R138Q Pathological Variant" International Journal of Molecular Sciences 23, no. 20: 12178. https://doi.org/10.3390/ijms232012178






