Effect of Heading Date on the Starch Structure and Grain Yield of Rice Lines with Low Gelatinization Temperature
Abstract
:1. Introduction
2. Results
2.1. Nucleotide Sequence of Hd1 in Kinmaze, Akita 63, and Akitakomachi
2.2. Genotyping and Western Blotting of Rice Accessions with Different Hd1 and SSIIa Alleles
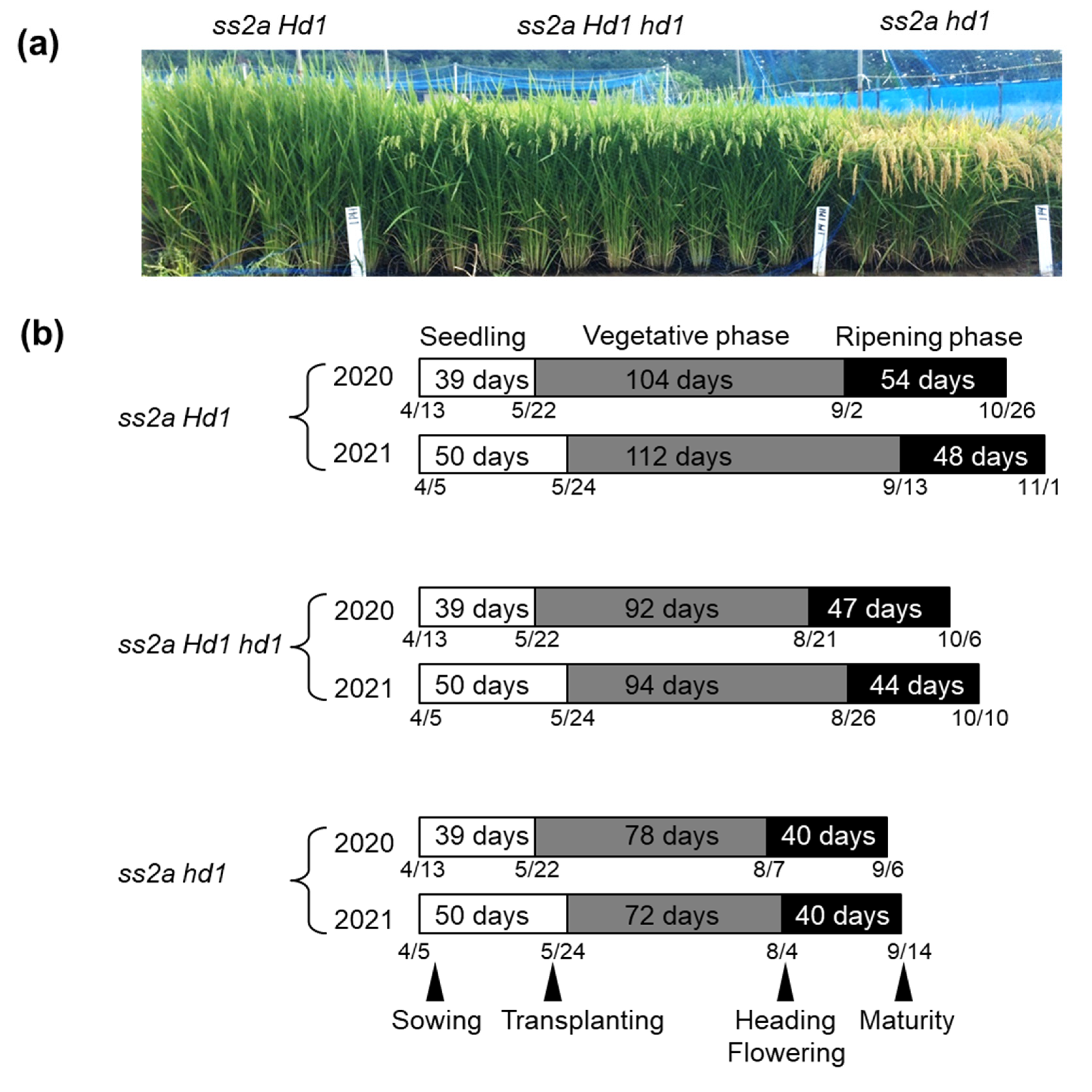
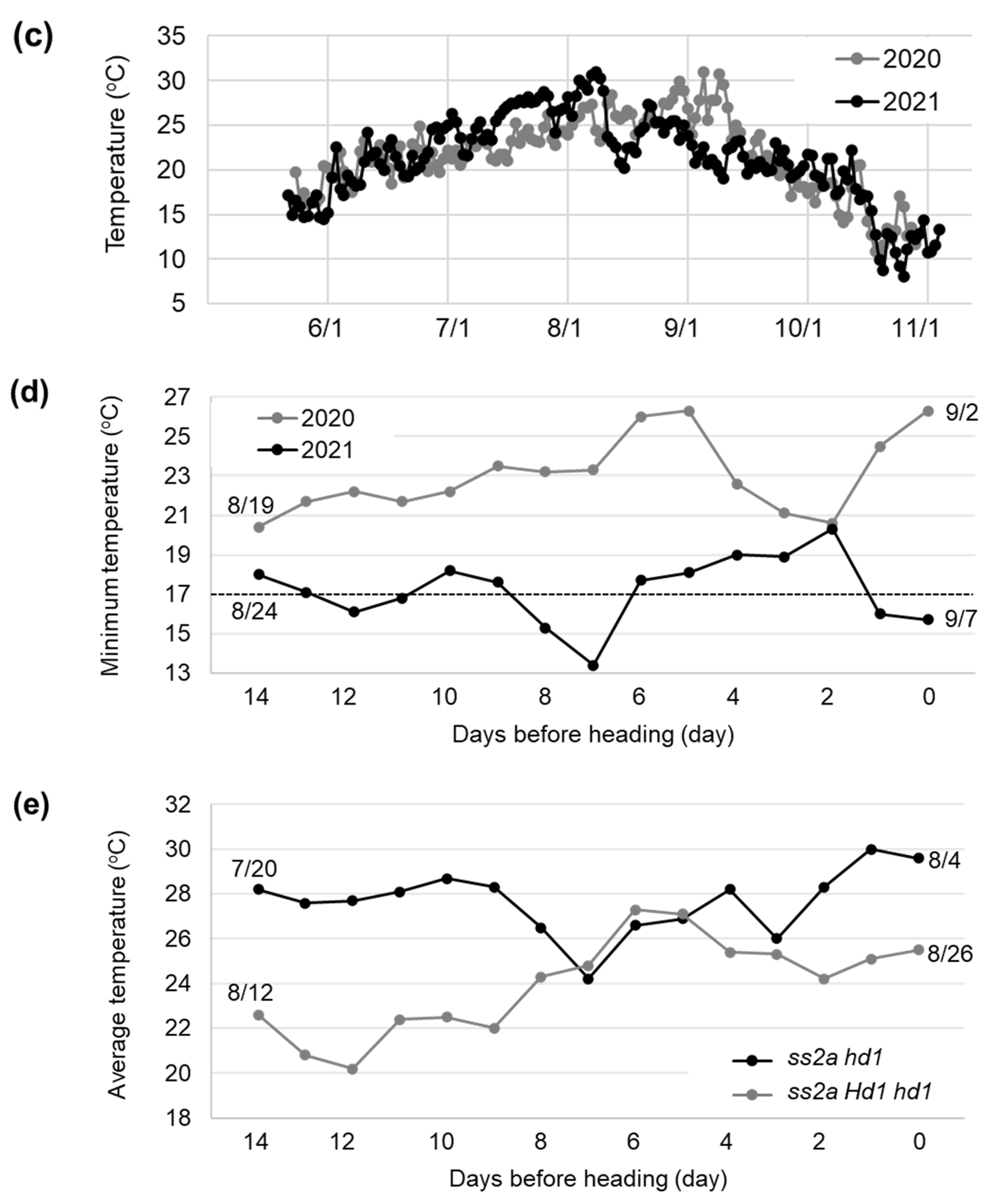
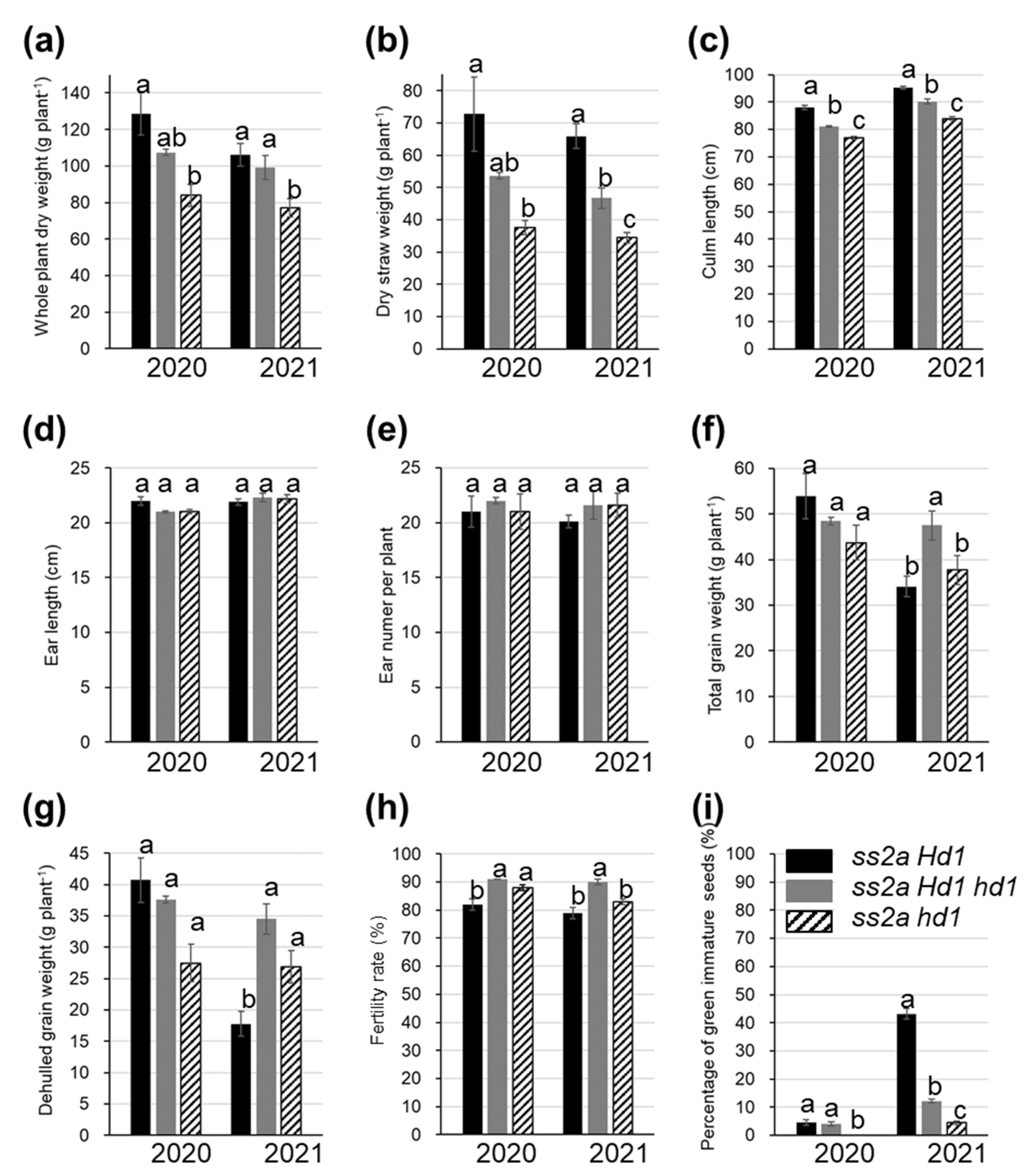
2.3. Effect of Hd1 Alleles on the Agricultural Traits of NILs
2.4. Effect of Hd1 Alleles on Apparent Amylose Content and GBSSI Expression Level
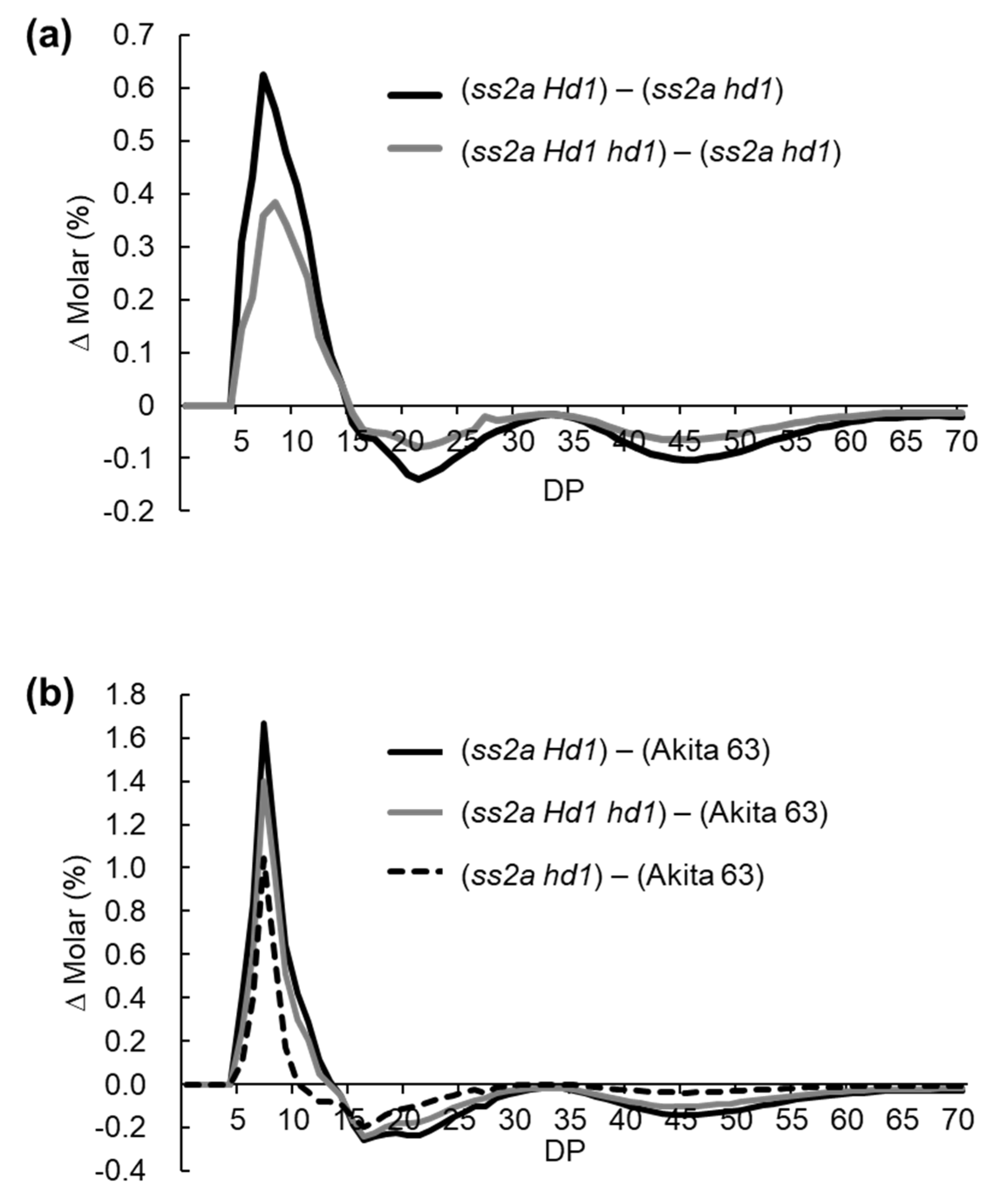
2.5. Effect of Hd1 Alleles on Amylopectin Structure
2.6. Effect of Hd1 Alleles on the Thermal Properties of Starch
3. Discussion
3.1. SNPs in Hd1
3.2. Effect of Hd1 Alleles on Grain Yield
3.3. Effect of Hd1 Alleles on Starch Structure
3.4. Effect of Hd1 Alleles on Starch Gelatinization Temperature
4. Materials and Methods
4.1. Plant Materials
4.2. Sequencing of the Hd1 Gene
4.3. Genotyping of Hd1 and SS2a Alleles
4.4. Field Experiments and Agricultural Traits
4.5. Meteorological Data
4.6. Western Blot Analysis
4.7. Measurement of Apparent Amylose Content and Short to Long Chain Amylopectin Ratio
4.8. Analysis of Amylopectin Structure
4.9. Measurement of Gelatinization Temperature
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, C.C.; Wei, F.J.; Chiou, W.Y.; Tsai, Y.C.; Wu, H.P.; Gotarkar, D.; Wei, Z.H.; Lai, M.H.; Hsing, Y.C. Studies of rice Hd1 haplotypes worldwide reveal adaptation of flowering time to different environments. PLoS ONE 2020, 15, e0239028. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2007, 58, 3091–3097. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Torollo, G.; Yoon, M.R.; Kwak, J.; Lee, C.K.; Prahalada, G.D.; Choi, I.R.; Yeo, U.S.; Jeong, O.Y.; Jena, K.K.; et al. Loss-of-Function Alleles of Heading date 1 (Hd1) are associated with adaptation of temperate japonica rice plants to the tropical region. Front. Plant Sci. 2018, 9, 1827. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Matsubara, K.; Kono, I.; Hori, K.; Nonoue, Y.; Ono, N.; Shomura, A.; Mizubayashi, T.; Yamamoto, S.; Yamanouchi, U.; Shirasawa, K.; et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theor. Appl. Genet. 2008, 117, 935–945. [Google Scholar] [CrossRef]
- Nonoue, Y.; Fujino, K.; Hirayama, Y.; Yamanouchi, U.; Lin, S.Y.; Yano, M. Detection of quantitative trait loci controlling extremely early heading in rice. Theor. Appl. Genet. 2008, 116, 715–722. [Google Scholar] [CrossRef]
- Takahashi, Y.; Teshima, K.M.; Yokoi, S.; Innan, H.; Shimamoto, K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 2009, 106, 4555–4560. [Google Scholar] [CrossRef]
- Matsubara, K.; Yamanouchi, U.; Nonoue, Y.; Sugimoto, K.; Wang, Z.X.; Minobe, Y.; Yano, M. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011, 66, 603–612. [Google Scholar] [CrossRef]
- Taoka, K.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef]
- Nemoto, Y.; Nonoue, Y.; Yano, M.; Izawa, T. Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016, 86, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Matsubara, K.; Yano, M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 2016, 129, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Chen, Y.; Liu, L.; Wang, Y.; Bao, S.; Zhou, X.; Teo, Z.W.; Mao, C.; Gan, Y.; Yu, H. OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKγ4. Plant Cell 2017, 29, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Du, A.; Tian, W.; Wei, M.; Yan, W.; He, H.; Zhou, D.; Huang, X.; Li, S.; Ouyang, X. The DTH8-Hd1 Module Mediates Day-Length-Dependent Regulation of Rice Flowering. Mol. Plant. 2017, 10, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Peng, Q.; Fu, D.; Zhuang, D.; Yu, Y.; Duan, M.; Xie, W.; Cai, Y.; Ouyang, Y.; Lian, X.; et al. The E3 ubiquitin ligase HAF1 modulates circadian accumulation of EARLY FLOWERING3 to control heading date in rice under long-day conditions. Plant Cell 2018, 30, 2352–2367. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shimamoto, K. Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice. Genes Genet. Syst. 2011, 86, 175–182. [Google Scholar] [CrossRef]
- Mo, Y.; Lee, C.M.; Park, H.M.; Ha, S.K.; Kim, M.J.; Kwak, J.; Lee, H.S.; Lee, J.H.; Jeung, J.U. Hd1 allele types and their associations with major agronomic traits in Korean rice cultivars. Plants 2021, 10, 2408. [Google Scholar] [CrossRef]
- Hizukuri, S. Polymodal distribution of the chain lengths of amylopectins, and its significance. Carbohydr. Res. 1986, 147, 342–347. [Google Scholar] [CrossRef]
- Takeda, Y.; Hizukuri, S.; Takeda, C.; Suzuki, A. Structures of branched molecules of amyloses of various origins, and molar fractions of branched and unbranched molecules. Carbohydr. Res. 1987, 165, 139–145. [Google Scholar] [CrossRef]
- Jane, J.; Chen, Y.Y.; Lee, L.F.; Mcpherson, A.E.; Wong, K.S.; Radosavljevic, M.; Kasemsuwan, T. Effects of amylopectin branch chain length and amylose content on the gelatinization and pasting properties of starch. Cereal. Chem. 1999, 76, 629–637. [Google Scholar] [CrossRef]
- Tanaka, N.; Fujita, N.; Nishi, A.; Satoh, H.; Hosaka, Y.; Ugaki, M.; Kawasaki, S.; Nakamura, Y. The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol. J. 2004, 2, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lin, Q.; Li, X.; Wang, F.; Chen, Z.; Wang, J.; Li, W.; Fan, F.; Tao, Y.; Jiang, Y.; et al. Fine-tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 2021, 19, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Sano, Y. Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 1984, 68, 467–473. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zheng, F.Q.; Shen, G.Z.; Gao, J.P.; Snustad, D.P.; Li, M.G.; Zhang, J.L.; Hong, M.M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995, 7, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.L.; Wang, Z.Y.; Xing, Y.Y.; Zhang, J.L.; Hong, M.M. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 1998, 14, 459–465. [Google Scholar] [CrossRef]
- Isshiki, M.; Morino, K.; Nakajima, M.; Okagaki, R.J.; Wessler, S.R.; Izawa, T.; Shimamoto, K.A. Naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 1998, 15, 133–138. [Google Scholar] [CrossRef]
- Hirano, H.Y.; Sano, Y. Enhancement of Wx gene expression and the accumulation of amylose in response to cool temperatures during seed development in rice. Plant Cell Physiol. 1998, 39, 807–812. [Google Scholar] [CrossRef]
- Larkin, P.D.; Park, W.D. Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 1999, 40, 719–727. [Google Scholar] [CrossRef]
- Zhang, H.; Duan, L.; Dai, J.S.; Zhang, C.Q.; Li, J.; Gu, M.H.; Liu, Q.Q.; Zhu, Y. Major QTLs reduce the deleterious effects of high temperature on rice amylose content by increasing splicing efficiency of Wx pre-mRNA. Theor. Appl. Genet. 2014, 27, 273–282. [Google Scholar] [CrossRef]
- Fujita, N. Starch biosynthesis in rice endosperm. AGri-Biosci. Monogr. 2014, 4, 1–18. [Google Scholar] [CrossRef]
- Crofts, N.; Nakamura, Y.; Fujita, N. Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci. 2017, 262, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Francisco, P.B., Jr.; Hosaka, Y.; Sato, A.; Sawada, T.; Kubo, A.; Fujita, N. Essential amino acids of starch synthase IIa differentiate amylopectin structure and starch quality between japonica and indica rice varieties. Plant Mol. Biol. 2005, 58, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: Rice endosperm as a model tissue. Plant Cell Physiol. 2002, 43, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Nishi, A.; Nakamura, Y.; Tanaka, N.; Satoh, H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 2001, 127, 459–472. [Google Scholar] [CrossRef]
- Butardo, V.M.; Fitzgerald, M.A.; Bird, A.R.; Gidley, M.J.; Flanagan, B.M.; Larroque, O.; Resurreccion, A.P.; Laidlaw, H.K.; Jobling, S.A.; Morell, M.K.; et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 2011, .62, 4927–4941. [Google Scholar] [CrossRef] [PubMed]
- Nakata, M.; Miyashita, T.; Kimura, R.; Nakata, Y.; Takagi, H.; Kuroda, M.; Yamaguchi, T.; Umemoto, T.; Yamakawa, H. MutMapPlus identified novel mutant alleles of a rice starch branching enzyme IIb gene for fine-tuning of cooked rice texture. Plant Biotechnol. J. 2018, 16, 111–123. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [CrossRef]
- Jiang, H.; Dian, W.; Wu, P. Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 2003, 63, 53–59. [Google Scholar] [CrossRef]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef] [Green Version]
- Ohdan, T.; Sawada, T.; Nakamura, Y. Effects of temperature on starch branching enzyme properties of rice. J. Appl. Glycosci. 2011, 58, 19–26. [Google Scholar] [CrossRef]
- Kato, K.; Suzuki, Y.; Hosaka, Y.; Takahashi, R.; Kodama, I.; Sato, K.; Kawamoto, T.; Kumamaru, T.; Fujita, N. Effect of high temperature on starch biosynthetic enzymes and starch structure in japonica rice cultivar ‘Akitakomachi’ (Oryza sativa L.) endosperm and palatability of cooked rice. J. Cereal Sci. 2019, 87, 209–214. [Google Scholar] [CrossRef]
- Miura, S.; Crofts, N.; Saito, Y.; Hosaka, Y.; Oitome, N.F.; Watanabe, T.; Kumamaru, T.; Fujita, N. Starch synthase IIa-deficient mutant rice line produces endosperm starch with lower gelatinization temperature than Japonica rice cultivars. Front. Plant Sci. 2018, 9, 645. [Google Scholar] [CrossRef]
- Makino, A.; Kaneta, Y.; Obara, M.; Ishiyama, K.; Kanno, K.; Kondo, E.; Suzuki, Y.; Mae, T. High yielding ability of a large-grain rice cultivar, Akita 63. Sci. Rep. 2020, 10, 12231. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y. Developing Isogenic Lines of Japanese Rice Cultivar ‘Koshihikari’ with Early and Late Heading. JARQ 2011, 45, 15–22. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Ando, I.; Nemoto, H.; Kato, H.; Hirabayashi, H.; Ohta, H.; Ishii, T.; Maeda, H.; Takemoto-kuno, Y.; Imbe, T.; et al. Breeding of “Milky Summer”, an isogenic line of the rice cultivar “Milky Queen” with modified heading. Bull. NARO Inst. Crop. Sci. 2013, 14, 77–95. [Google Scholar]
- Shibata, M. Present conditions and subjects of rice breeding for cold-tolerance in Japan. JARQ 1970, 5, 4. [Google Scholar]
- Satake, T. Determination of the most sensitive stage to sterile-type cool injury in rice plants. Res. Bull. Hokkaido Natl. Agric. Exp. Stn. 1976, 113, 1–43. [Google Scholar]
- Endo, M.; Tsuchiya, T.; Hamada, K.; Kawamura, S.; Yano, K.; Ohshima, M.; Higashitani, A.; Watanabe, M.; Kawagishi-Kobayashi, M. High temperatures cause male sterility in rice plants with transcriptional alterations during pollen development. Plant Cell Physiol. 2009, 50, 1911–1922. [Google Scholar] [CrossRef]
- Zhou, L.J.; Sheng, W.T.; Wu, J.; Zhang, C.Q.; Liu, Q.q.; Deng, Q.Y. Differential expressions among five Waxy alleles and their effects on the eating and cooking qualities in specialty rice cultivars. J. Integr. Agric. 2015, 14, 1153–1162. [Google Scholar] [CrossRef]
- Shi, S.; Wang, E.; Li, C.; Cai, M.; Cheng, B.; Cao, C.; Jiang, Y. Use of protein content, amylose content, and RVA parameters to evaluate the taste quality of rice. Front. Nutr. 2022, 13, 8. [Google Scholar] [CrossRef]
- Miura, S.; Koyama, N.; Crofts, N.; Hosaka, Y.; Abe, M.; Fujita, N. Generation and starch characterization of non-transgenic BEI and BEIIb double mutant rice (Oryza sativa) with ultra-high level of resistant starch. Rice 2021, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N. Manipulation of rice starch properties for application. In Starch; Nakamura, Y., Ed.; Springer: Tokyo, Japan, 2015; pp. 335–369. [Google Scholar]
- Hayashi, M.; Kodama, M.; Nakamura, Y.; Fujita, N. Thermal and pasting properties, morphology of starch granules, and crystallinity of endosperm starch in the rice SSI and SSIIIa double-mutant. J. Appl. Glycosci. 2015, 62, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Tsuiki, K.; Fujisawa, H.; Itoh, A.; Sato, M.; Fujita, N. Alterations of starch structure lead to increased resistant starch of steamed rice: Identification of high resistant starch rice lines. J. Cereal Sci. 2016, 68, 88–92. [Google Scholar] [CrossRef]
- Nagamatsu, S.; Wada, T.; Matsushima, R.; Fujita, N.; Miura, S.; Crofts, N.; Hosaka, Y.; Yamaguchi, O.; Kumamaru, T. Mutation in BEIIb mitigates the negative effect of the mutation in ISA1 on grain filling and amyloplast formation in rice. Plant Mol. Biol. 2022, 108, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jin, M.; Zheng, X.M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef]
- Lee, S.J.; Kang, K.; Lim, J.H.; Paek, N.C. Natural alleles of CIRCADIAN CLOCK ASSOCIATED1 contribute to rice cultivation by fine-tuning flowering time. Plant Physiol. 2022, 190, 640–656. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Asakura, N.; Ohdan, T.; Miyao, A.; Hirochika, H.; Nakamura, Y. Function and characterization of starch synthase I using mutants in rice. Plant Physiol. 2006, 140, 1070–1084. [Google Scholar] [CrossRef]
- Crofts, N.; Abe, N.; Oitome, N.F.; Matsushima, R.; Hayashi, M.; Tetlow, I.J.; Emes, M.J.; Nakamura, Y.; Fujita, N. Amylopectin biosynthetic enzymes from developing rice seed form enzymatically active protein complexes. J. Exp. Bot. 2015, 66, 4469–4482. [Google Scholar] [CrossRef]
- Yamamoto, K.; Sawada, S.; Onogaki, T. Properties of rice prepared by alkali method with various conditions. J. Jpn. Soc. Starch. Sci. 1973, 20, 99–104. [Google Scholar] [CrossRef]
- Yamamoto, K.; Sawada, S.; Onogaki, T. Effects of quality and quantity of alkaline solution on the properties of the rice starch. J. Jap. Soc. Starch Sci. 1981, 28, 241–244. [Google Scholar] [CrossRef]
- Horibata, T.; Nakamoto, M.; Fuwa, H.; Inouchi, N. Structural and physicochemical characteristics of endosperm starches of rice cultivars recently bred in Japan. J. Appl. Glycosci. 2004, 51, 303–313. [Google Scholar] [CrossRef]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, Y.; Nishi, A.; Satoh, H.; Park, J.H.; Jane, J.L.; et al. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toyosawa, Y.; Kawagoe, Y.; Matsushima, R.; Crofts, N.; Ogawa, M.; Fukuda, M.; Kumamaru, T.; Okazaki, Y.; Kusano, M.; Saito, K.; et al. Deficiency of starch synthase IIIa and IVb alters starch granule morphology from polyhedral to spherical in rice endosperm. Plant Physiol. 2016, 170, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Hasegawa, H.; Taira, T. The isolation and characterization of waxy mutant of diploid wheat (Triticum monococcum L.). Plant Sci. 2001, 160, 595–602. [Google Scholar] [CrossRef]
- Fujita, N.; Kubo, A.; Suh, D.; Wong, K.; Jane, J.; Ozawa, K.; Takaiwa, F.; Inaba, Y.; Nakamura, Y. Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol. 2003, 44, 607–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Rice Accession | SSIIa Genotype 1 | Hd1 Genotype | Heading Date 2 |
|---|---|---|---|
| Nipponbare | ss2aL | Hd1 | Late August |
| Kasalath | SS2a | hd1Kas | Early August |
| Akitakomachi | ss2aL | hd1 | Late July |
| Akita 63 | ss2aL | hd1 | Early August |
| Kinmaze | ss2aL | Hd1 | Early September |
| EM204 | ss2a | Hd1 | Early September |
| ss2a Hd1 | ss2a | Hd1 | Early September |
| ss2a Hd1 hd1 | ss2a | Hd1 hd1 | Late August |
| ss2a hd1 | ss2a | hd1 | Early August |
| Rice Accession | Apparent Amylose Content (%) 1 | Ratio of Short to Long Chains of Amylopectin 1 | ||
|---|---|---|---|---|
| 2020 | 2021 | 2020 | 2021 | |
| Akita 63 | 18.1 ± 0.8 | 17.1 ± 0.4 | 2.4 ± 0.1 | 2.7 ± 0.1 |
| ss2a Hd1 | 27.0 ± 0.6 a* | 28.0 ± 0.1 a* | 3.1 ± 0.0 a* | 3.2 ± 0.0 a* |
| ss2a Hd1 hd1 | 25.1 ± 0.6 ab* | 26.6 ± 0.6 ab* | 2.6 ± 0.0 b | 2.9 ± 0.0 b |
| ss2a hd1 | 22.1 ± 0.4 b* | 24.7 ± 0.6 b* | 2.4 ± 0.0 b | 3.0 ± 0.0 ab |
| Rice Accession | Tp (°C) 1 | |
|---|---|---|
| 2020 | 2021 | |
| Akita 63 | 63.1 ± 0.0 a | 62.0 ± 0.1 a |
| ss2a Hd1 | 50.3 ± 0.1 d | 52.3 ± 0.2 d |
| ss2a Hd1 hd1 | 58.8 ± 0.1 c | 55.5 ± 0.1 c |
| ss2a hd1 | 61.8 ± 0.1 b | 57.2 ± 0.1 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crofts, N.; Hareyama, K.; Miura, S.; Hosaka, Y.; Oitome, N.F.; Fujita, N. Effect of Heading Date on the Starch Structure and Grain Yield of Rice Lines with Low Gelatinization Temperature. Int. J. Mol. Sci. 2022, 23, 10783. https://doi.org/10.3390/ijms231810783
Crofts N, Hareyama K, Miura S, Hosaka Y, Oitome NF, Fujita N. Effect of Heading Date on the Starch Structure and Grain Yield of Rice Lines with Low Gelatinization Temperature. International Journal of Molecular Sciences. 2022; 23(18):10783. https://doi.org/10.3390/ijms231810783
Chicago/Turabian StyleCrofts, Naoko, Kaito Hareyama, Satoko Miura, Yuko Hosaka, Naoko F. Oitome, and Naoko Fujita. 2022. "Effect of Heading Date on the Starch Structure and Grain Yield of Rice Lines with Low Gelatinization Temperature" International Journal of Molecular Sciences 23, no. 18: 10783. https://doi.org/10.3390/ijms231810783




