Role of the Cysteine in R3 Tau Peptide in Copper Binding and Reactivity
Abstract
:1. Introduction
2. Results and Discussion
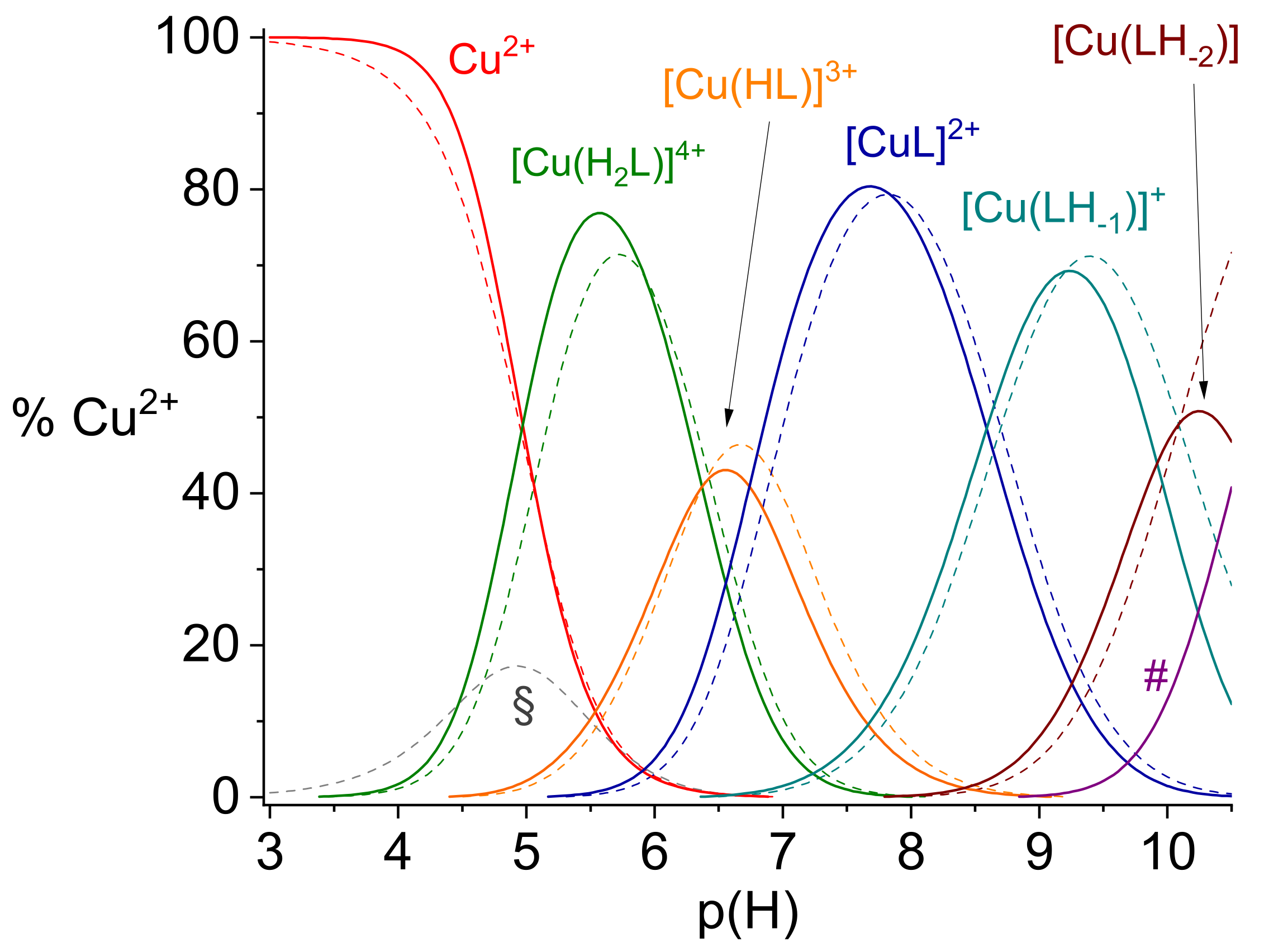
2.1. Protonation and Copper(II) Complexation Equilibria
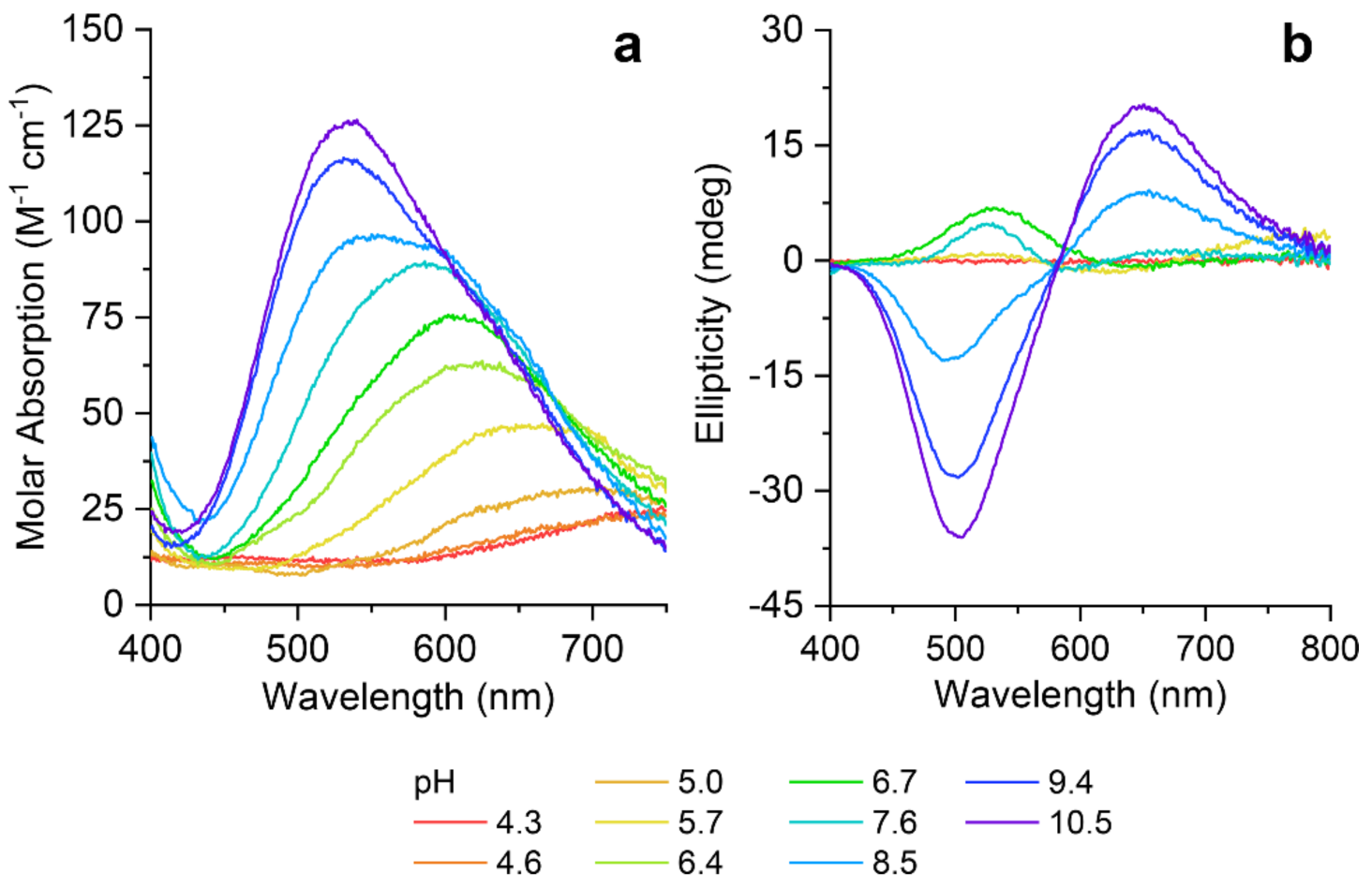
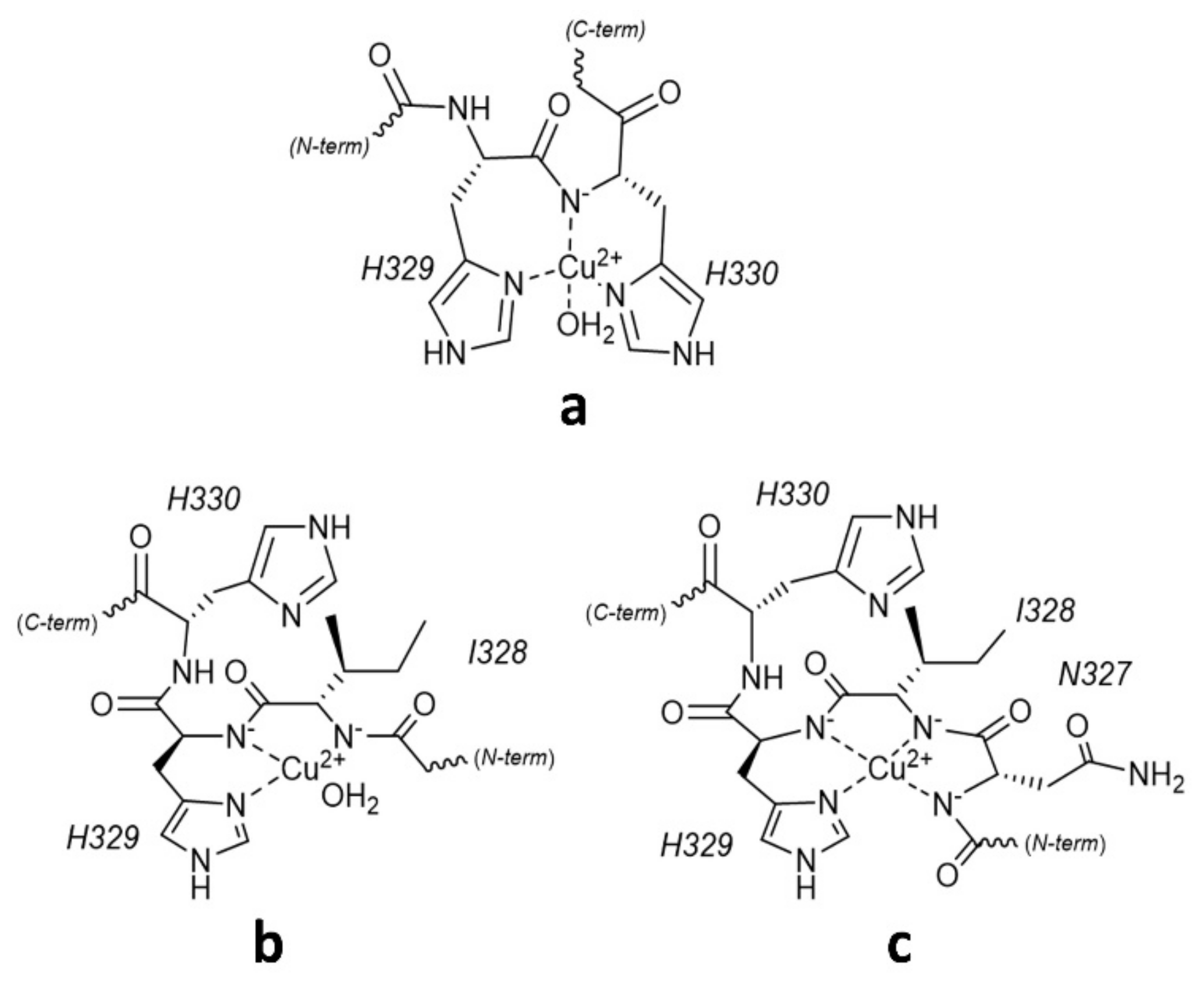
2.2. Copper(II) Complexation Studies
Cu2+/R3A Complex Formation Equilibria
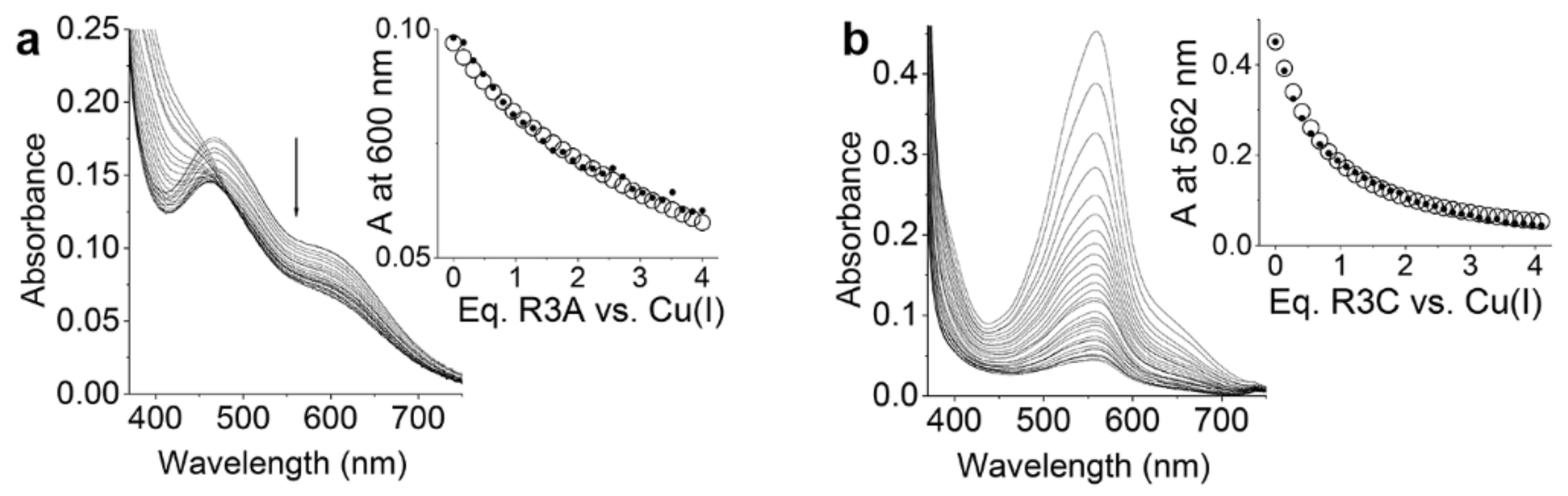
2.3. Cu+/R3 and R3A Complex Formation Equilibria
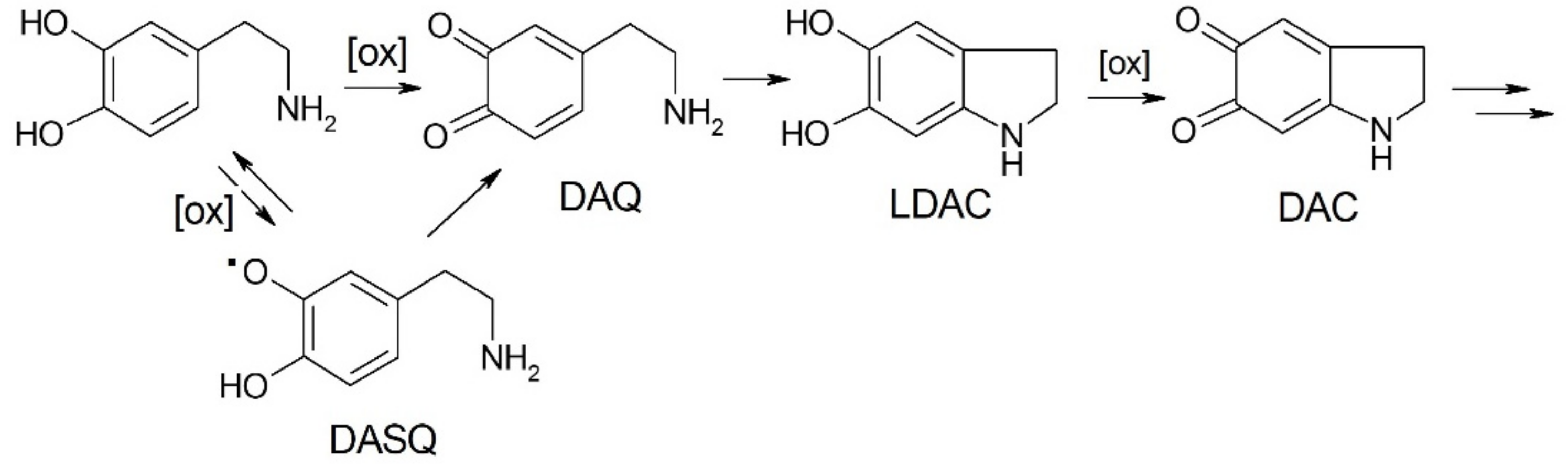
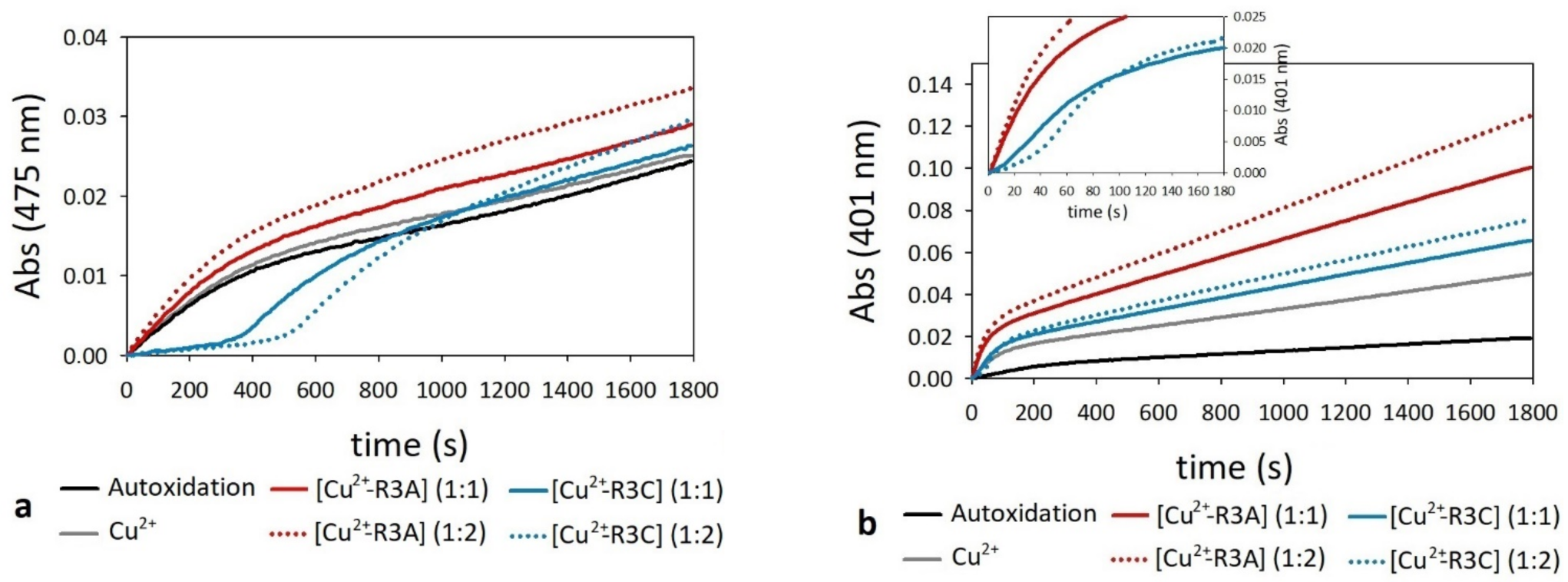
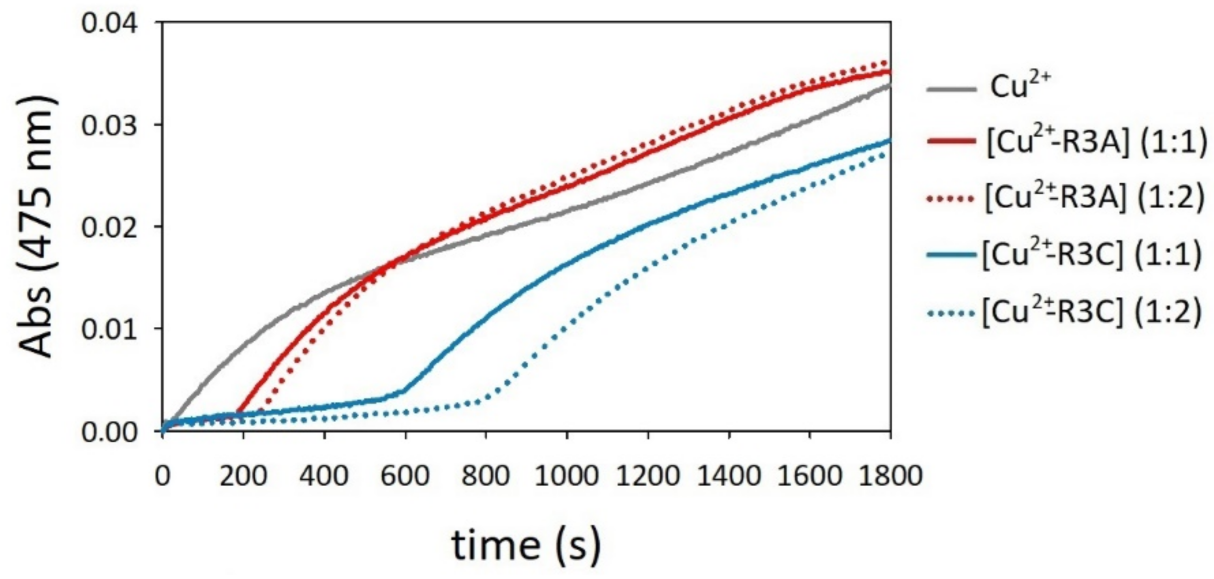
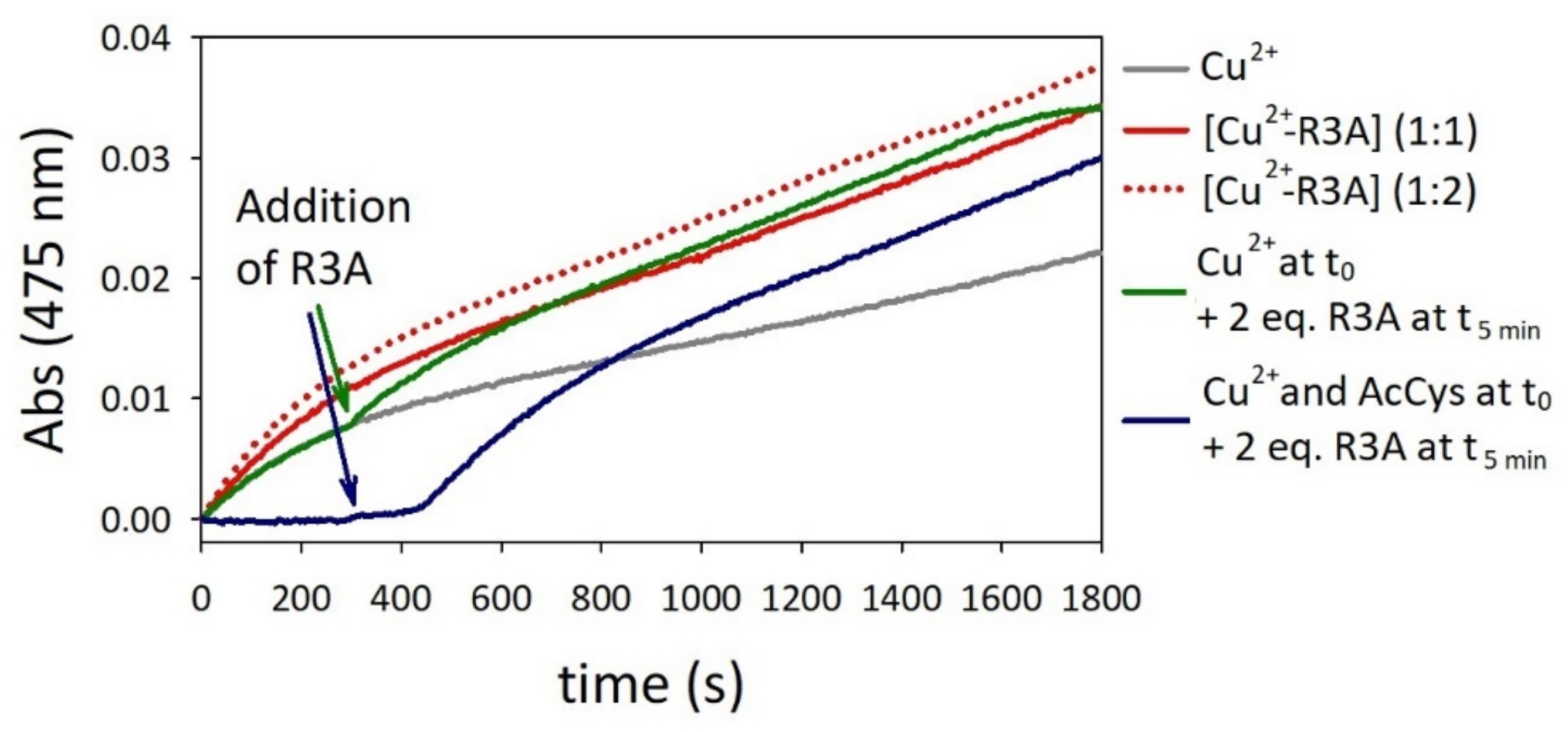
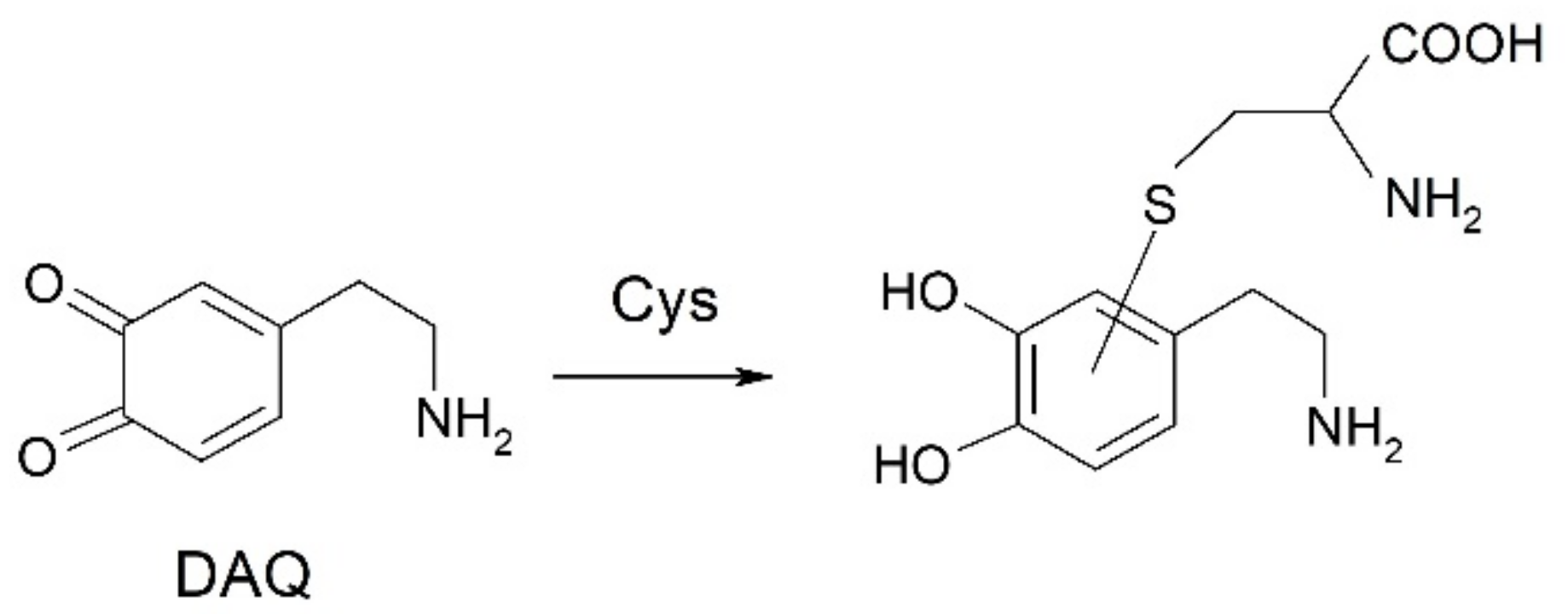
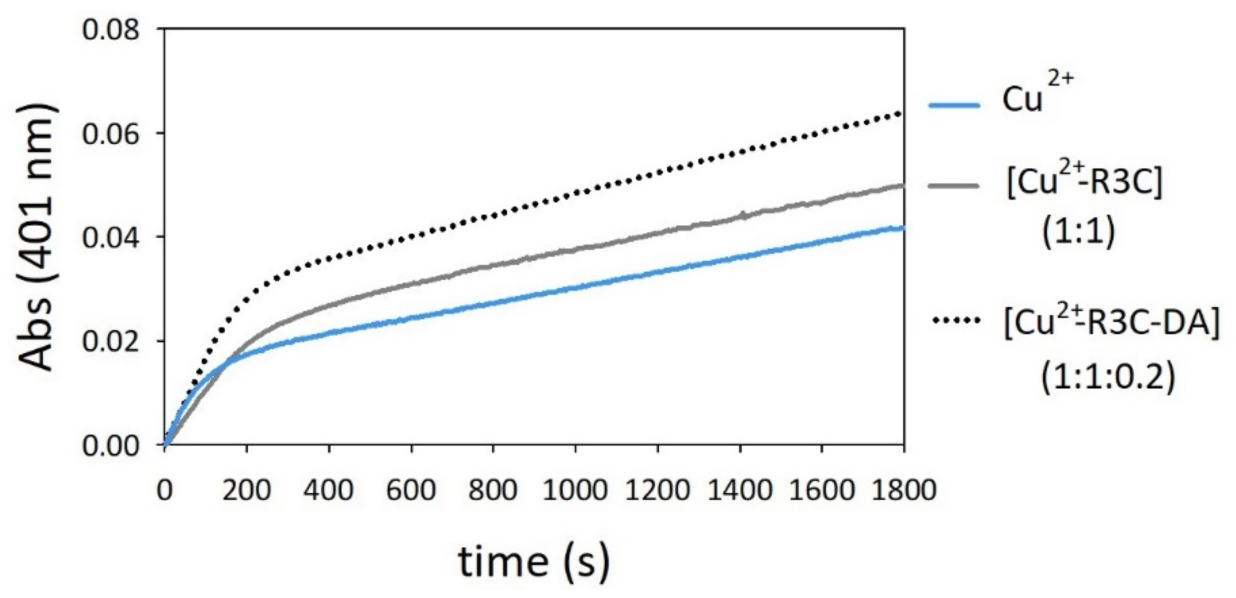
2.4. Oxidation of Dopamine and 4-Methylcatechol by Copper–R3C and Copper–R3A Complexes
- Cu2+ + peptide ⇄ [Cu2+-peptide]
- [Cu2+-peptide] + catechol → [Cu+-peptide] + semiquinone•+
- [Cu+-peptide] + catechol ⇄ [Cu+-peptide-catechol]
- [Cu+-peptide-catechol] + O2 → [Cu-peptide-catechol-O2]
- [Cu-peptide-catechol-O2] →→ [Cu2+-peptide] + quinone
- 2 semiquinone•+ → catechol + quinone
- 7.
- [Cu2+-peptide] + R3C → [Cu+-peptide] + R3C-S• + H+
- 8.
- [Cu+-peptide] + O2 → [Cu-peptide-O2]
- 9.
- [Cu-peptide-O2] + R3C → [Cu2+-peptide] + R3C-S• + H+
- 10.
- DA →→ →DASQ•
- 11.
- Cys-SH + DASQ• → Cys-S• + DA
- 12.
- 2 Cys-S• → Cys-S-S-Cys
2.5. Competitive Endogenous R3C and R3A Peptide Oxidation
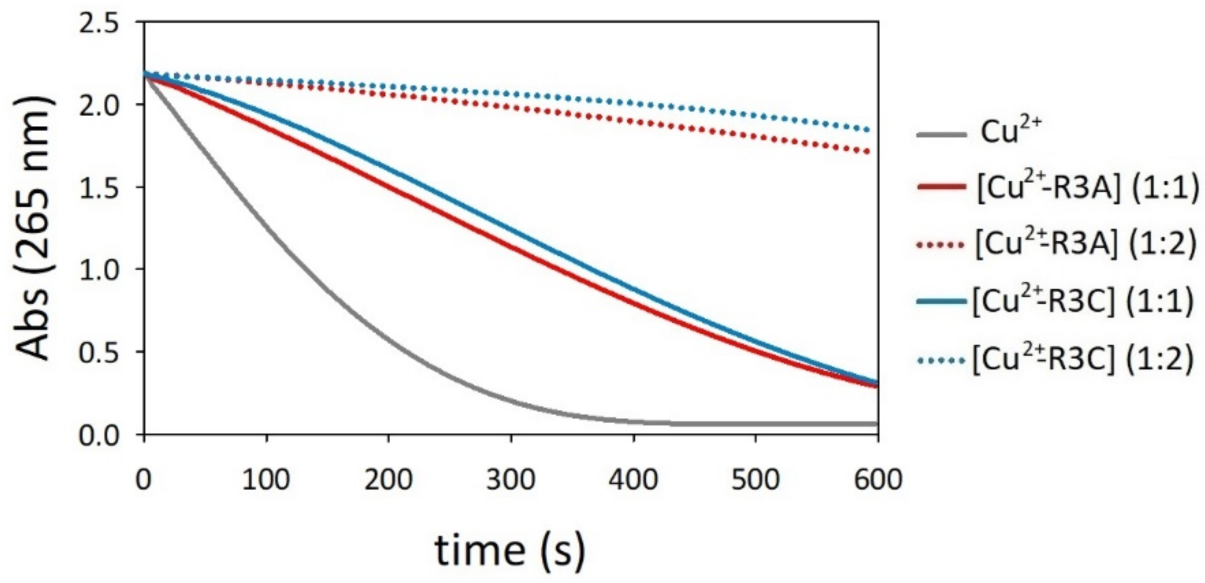
2.6. Oxidation of Ascorbate by Copper–R3C and Copper–R3A Complexes
3. Materials and Methods
3.1. Materials and Instrumentation
3.2. Peptide Synthesis
3.3. Complex Formation Equilibria Studies
3.4. Potentiometric Measurements
3.5. UV-Visible Absorption and Circular Dichroism Studies
3.6. Kinetics of Oxidation of Catechols and Ascorbate in the Presence of [Cu-R3A] and [Cu-R3C] Complexes
3.7. Kinetics of Oxidation of Catechols in the Presence of Cu and N-Acetylcysteine/Cystine
3.8. Identification and Characterization of Modified Peptides by HPLC-ESI/MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Sanabria-Castro, A.; Alvarado-Echeverría, I.; Monge-Bonilla, C. Molecular Pathogenesis of Alzheimer’s Disease: An Update. Ann. Neurosci. 2017, 24, 46–54. [Google Scholar] [CrossRef]
- Wang, Y.; Mandelkow, E. Tau in physiology and pathology. Nat. Rev. Neurosci. 2016, 17, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Noble, W.; Hanger, D.P. Roles of tau protein in health and disease. Acta Neuropathol. 2017, 133, 665–704. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Scheres, S.H.; Zhang, W.; Falcon, B.; Goedert, M. Cryo-EM structures of tau filaments. Curr. Opin. Struct. Biol. 2020, 64, 17–25. [Google Scholar] [CrossRef]
- Kidd, M. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature 1963, 197, 192–193. [Google Scholar] [CrossRef]
- Noble, W.; Hanger, D.P.; Miller, C.C.; Lovestone, S. The importance of tau phosphorylation for neurodegenerative diseases. Front. Neurol. 2013, 4, 83. [Google Scholar] [CrossRef]
- Brister, M.A.; Pandey, A.K.; Bielska, A.A.; Zondlo, N.J. OGlcNAcylation and Phosphorylation Have Opposing Structural Effects in tau: Phosphothreonine Induces Particular Conformational Order. J. Am. Chem. Soc. 2014, 136, 3803–3816. [Google Scholar] [CrossRef]
- Wang, J.Z.; Grundke-Iqbal, I.; Iqbal, K. Kinases and phosphatases and tau sites involved in Alzheimer neurofibrillary degeneration. Eur. J. Neurosci. 2007, 25, 59–68. [Google Scholar] [CrossRef]
- Iqbal, K.; Grundke-Iqbal, I. Alzheimer neurofibrillary degeneration: Significance, etiopathogenesis, therapeutics and prevention. J. Cell Mol. Med. 2008, 12, 38–55. [Google Scholar] [CrossRef]
- Lippens, G.; Gigant, B. Elucidating Tau function and dysfunction in the era of cryo-EM. J. Biol. Chem. 2019, 294, 9316–9325. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Falcon, B.; He, S.; Murzin, A.G.; Murshudov, G.; Garringer, H.J.; Crowther, R.A.; Ghetti, B.; Goedert, M.; Scheres, S.H.W. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 2017, 547, 185. [Google Scholar] [CrossRef] [PubMed]
- Pradeepkiran, J.A.; Reddy, P.H. Structure Based Design and Molecular Docking Studies for Phosphorylated Tau Inhibitors in Alzheimer’s Disease. Cells 2019, 8, 260. [Google Scholar] [CrossRef] [PubMed]
- Pradeepkiran, J.A.; Munikumar, M.; Reddy, A.P.; Reddy, P.H. Protective effects of a small molecule inhibitor ligand against hyperphosphorylated tau-induced mitochondrial and synaptic toxicities in Alzheimer disease. Hum. Mol. Genet. 2021, 31, 244–261. [Google Scholar] [CrossRef]
- Cohen, T.J.; Friedmann, D.; Hwang, A.W.; Marmorstein, R.; Lee, V.M. The microtubule-associated tau protein has intrinsic acetyltransferase activity. Nat. Struct. Mol. Biol 2013, 20, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Martinho, M.; Allegro, D.; Huvent, I.; Chabaud, C.; Etienne, E.; Kovacic, H.; Guigliarelli, B.; Peyrot, V.; Landrieu, I.; Belle, V.; et al. Two Tau binding sites on tubulin revealed by thiol-disulfide exchanges. Sci. Rep. 2018, 8, 13846. [Google Scholar] [CrossRef]
- Barbier, P.; Zejneli, O.; Martinho, M.; Lasorsa, A.; Belle, V.; Smet-Nocca, C.; Tsvetkov, P.O.; Devred, F.; Landrieu, I. Role of Tau as a Microtubule-Associated Protein: Structural and Functional Aspects. Front. Aging Neurosci. 2019, 11, 204. [Google Scholar] [CrossRef]
- Furukawa, Y.; Kaneko, K.; Nukina, N. Tau protein assembles into isoform- and disulfide-dependent polymorphic fibrils with distinct structural properties. J. Biol. Chem. 2011, 286, 27236–27246. [Google Scholar] [CrossRef]
- Chidambaram, H.; Chinnathambi, S. Role of cysteines in accelerating Tau filament formation. J. Biomol. Struct. Dyn. 2020, 40, 4366–4375. [Google Scholar] [CrossRef]
- Bhattacharya, K.; Rank, K.B.; Evans, D.B.; Sharma, S.K. Role of cysteine-291 and cysteine-322 in the polymerization of human tau into Alzheimer-like filaments. Biochem. Biophys. Res. Commun. 2001, 285, 20–26. [Google Scholar] [CrossRef]
- Soeda, Y.; Yoshikawa, M.; Almeida, O.F.; Sumioka, A.; Maeda, S.; Osada, H.; Kondoh, Y.; Saito, A.; Miyasaka, T.; Kimura, T.; et al. Toxic tau oligomer formation blocked by capping of cysteine residues with 1,2-dihydroxybenzene groups. Nat. Commun. 2015, 6, 10216. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lim, S.; Haque, M.M.; Ryoo, N.; Hong, H.S.; Rhim, H.; Lee, D.E.; Chang, Y.T.; Lee, J.S.; Cheong, E.; et al. Identification of disulfide cross-linked tau dimer responsible for tau propagation. Sci. Rep. 2015, 5, 15231. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.; Ullman, O.; Stultz, C.M. Using intramolecular disulfide bonds in tau protein to deduce structural features of aggregation-resistant conformations. J. Biol. Chem. 2012, 287, 9591–9600. [Google Scholar] [CrossRef] [PubMed]
- Barghorn, S.; Mandelkow, E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry 2002, 41, 14885–14896. [Google Scholar] [CrossRef]
- Schweers, O.; Mandelkow, E.M.; Biernat, J.; Mandelkow, E. Oxidation of cysteine-322 in the repeat domain of microtubule-associated protein tau controls the in vitro assembly of paired helical filaments. Proc. Natl. Acad. Sci. USA 1995, 92, 8463–8467. [Google Scholar] [CrossRef] [PubMed]
- Al-Hilaly, Y.K.; Pollack, S.J.; Vadukul, D.M.; Citossi, F.; Rickard, J.E.; Simpson, M.; Storey, J.M.D.; Harrington, C.R.; Wischik, C.M.; Serpell, L.C. Alzheimer’s Disease-like Paired Helical Filament Assembly from Truncated Tau Protein Is Independent of Disulfide Crosslinking. J. Mol. Biol. 2017, 429, 3650–3665. [Google Scholar] [CrossRef]
- Prifti, E.; Tsakiri, E.N.; Vourkou, E.; Stamatakis, G.; Samiotaki, M.; Papanikolopoulou, K. The two cysteines of tau protein are functionally distinct and contribute differentially to its pathogenicity in vivo. J. Neurosci. 2021, 41, 797–810. [Google Scholar] [CrossRef]
- Lei, P.; Ayton, S.; Bush, A.I. The Essential Elements of Alzheimer’s Disease. J. Biol. Chem. 2020, 296, 100105. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Boyd-Kimball, D. Oxidative Stress, Amyloid-β Peptide, and Altered Key Molecular Pathways in the Pathogenesis and Progression of Alzheimer’s Disease. J. Alzheimers Dis 2018, 62, 1345–1367. [Google Scholar] [CrossRef]
- Scheiber, I.F.; Mercer, J.F.; Dringen, R. Metabolism and functions of copper in brain. Prog. Neurobiol. 2014, 116, 33–57. [Google Scholar] [CrossRef]
- Chang, C.J. Bioinorganic Life and Neural Activity: Toward a Chemistry of Consciousness? Acc. Chem. Res. 2017, 50, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Sensi, S.L.; Granzotto, A.; Siotto, M.; Squitti, R. Copper and Zinc Dysregulation in Alzheimer’s Disease. Trends Pharmacol. Sci. 2018, 39, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Kepp, K.P.; Squitti, R. Copper imbalance in Alzheimer’s disease: Convergence of the chemistry and the clinic. Coord. Chem. Rev. 2019, 397, 168–187. [Google Scholar] [CrossRef]
- Zubčić, K.; Hof, P.R.; Šimić, G.; Jazvinšćak Jembrek, M. The Role of Copper in Tau-Related Pathology in Alzheimer’s Disease. Front. Mol. Neurosci. 2020, 13, 572308. [Google Scholar] [CrossRef] [PubMed]
- Harris, C.J.; Gray, N.E.; Caruso, M.; Hunter, M.; Ralle, M.; Quinn, J.F. Copper Modulation and Memory Impairment due to Hippocampal Tau Pathology. J. Alzheimer’s Dis. 2020, 78, 49–60. [Google Scholar] [CrossRef]
- Ahmadi, S.; Zhu, S.; Sharma, R.; Wilson, D.J.; Kraatz, H.B. Interaction of metal ions with tau protein. The case for a metal-mediated tau aggregation. J. Inorg. Biochem. 2019, 194, 44–51. [Google Scholar] [CrossRef]
- Ahmadi, S.; Zhu, S.; Sharma, R.; Wu, B.; Soong, R.; Dutta Majumdar, R.; Wilson, D.J.; Simpson, A.J.; Kraatz, H.B. Aggregation of Microtubule Binding Repeats of Tau Protein is Promoted by Cu2+. ACS Omega 2019, 4, 5356–5366. [Google Scholar] [CrossRef]
- Soragni, A.; Zambelli, B.; Mukrasch, M.D.; Biernat, J.; Jeganathan, S.; Griesinger, C.; Ciurli, S.; Mandelkow, E.; Zweckstetter, M. Structural Characterization of Binding of Cu(II) to Tau Protein†. Biochemistry 2008, 47, 10841–10851. [Google Scholar] [CrossRef]
- Zhou, L.X.; Du, J.T.; Zeng, Z.Y.; Wu, W.H.; Zhao, Y.F.; Kanazawa, K.; Ishizuka, Y.; Nemoto, T.; Nakanishi, H.; Li, Y.M. Copper (II) modulates in vitro aggregation of a tau peptide. Peptides 2007, 28, 2229–2234. [Google Scholar] [CrossRef]
- Ma, Q.-F.; Li, Y.-M.; Du, J.-T.; Kanazawa, K.; Nemoto, T.; Nakanishi, H.; Zhao, Y.-F. Binding of copper (II) ion to an Alzheimer’s tau peptide as revealed by MALDI-TOF MS, CD, and NMR. Biopolymers 2005, 79, 74–85. [Google Scholar] [CrossRef]
- Ma, Q.; Li, Y.; Du, J.; Liu, H.; Kanazawa, K.; Nemoto, T.; Nakanishi, H.; Zhao, Y. Copper binding properties of a tau peptide associated with Alzheimer’s disease studied by CD, NMR, and MALDI-TOF MS. Peptides 2006, 27, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.-k.; Saxena, S. Insight into Potential Cu(II)-Binding Motifs in the Four Pseudorepeats of Tau Protein. J. Phys. Chem. B 2011, 115, 15067–15078. [Google Scholar] [CrossRef] [PubMed]
- Balogh, B.D.; Szakács, B.; Di Natale, G.; Tabbì, G.; Pappalardo, G.; Sóvágó, I.; Várnagy, K. Copper (II) binding properties of an octapeptide fragment from the R3 region of tau protein: A combined potentiometric, spectroscopic and mass spectrometric study. J. Inorg. Biochem. 2021, 217, 111358. [Google Scholar] [CrossRef] [PubMed]
- Lukács, M.; Szunyog, G.; Grenács, Á.; Lihi, N.; Kállay, C.; Di Natale, G.; Campagna, T.; Lanza, V.; Tabbi, G.; Pappalardo, G.; et al. Copper(II) Coordination Abilities of the Tau Protein’s N-Terminus Peptide Fragments: A Combined Potentiometric, Spectroscopic and Mass Spectrometric Study. ChemPlusChem 2019, 84, 1697–1708. [Google Scholar] [CrossRef]
- Di Natale, G.; Bellia, F.; Sciacca, M.F.M.; Campagna, T.; Pappalardo, G. Tau-peptide fragments and their copper(II) complexes: Effects on Amyloid-β aggregation. Inorg. Chim. Acta 2018, 472, 82–92. [Google Scholar] [CrossRef]
- Jing, J.; Tu, G.; Yu, H.; Huang, R.; Ming, X.; Zhan, H.; Zhan, F.; Xue, W. Copper (Cu(2+)) ion-induced misfolding of tau protein R3 peptide revealed by enhanced molecular dynamics simulation. Phys. Chem. Chem. Phys. 2021, 23, 11717–11726. [Google Scholar] [CrossRef]
- Bacchella, C.; Gentili, S.; Bellotti, D.; Quartieri, E.; Draghi, S.; Baratto, M.C.; Remelli, M.; Valensin, D.; Monzani, E.; Nicolis, S.; et al. Binding and Reactivity of Copper to R1 and R3 Fragments of tau Protein. Inorg. Chem. 2020, 59, 274–286. [Google Scholar] [CrossRef]
- Chen, S.H.; Li, C.W. Detection and Characterization of Catechol Quinone-Derived Protein Adducts Using Biomolecular Mass Spectrometry. Front. Chem. 2019, 7, 571. [Google Scholar] [CrossRef]
- Yang, J.; Cohen Stuart, M.A.; Kamperman, M. Jack of all trades: Versatile catechol crosslinking mechanisms. Chem. Soc. Rev. 2014, 43, 8271–8298. [Google Scholar] [CrossRef]
- Alies, B.; Badei, B.; Faller, P.; Hureau, C. Reevaluation of copper(I) affinity for amyloid-beta peptides by competition with ferrozine—An unusual copper(I) indicator. Chemistry 2012, 18, 1161–1167. [Google Scholar] [CrossRef]
- Aquilanti, G.; Giorgetti, M.; Minicucci, M.; Papini, G.; Pellei, M.; Tegoni, M.; Trasatti, A.; Santini, C. A study on the coordinative versatility of new N,S-donor macrocyclic ligands: XAFS, and Cu2+ complexation thermodynamics in solution. Dalton Trans. 2011, 40, 2764–2777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, R.; Wang, G. Impact of Dopamine Oxidation on Dopaminergic Neurodegeneration. ACS Chem. Neurosci. 2019, 10, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Monzani, E.; Nicolis, S.; Dell’Acqua, S.; Capucciati, A.; Bacchella, C.; Zucca, F.A.; Mosharov, E.V.; Sulzer, D.; Zecca, L.; Casella, L. Dopamine, Oxidative Stress and Protein-Quinone Modifications in Parkinson’s and Other Neurodegenerative Diseases. Angew. Chem. Int. Ed. Engl. 2019, 58, 6512–6527. [Google Scholar] [CrossRef] [PubMed]
- Herlinger, E.; Jameson, R.F.; Linert, W. Spontaneous autoxidation of dopamine. J. Chem. Soc. Perkin Trans. 2 1995, 2, 259–263. [Google Scholar] [CrossRef]
- Dell’Acqua, S.; Pirota, V.; Anzani, C.; Rocco, M.M.; Nicolis, S.; Valensin, D.; Monzani, E.; Casella, L. Reactivity of copper-[small alpha]-synuclein peptide complexes relevant to Parkinson’s disease. Metallomics 2015, 7, 1091–1102. [Google Scholar] [CrossRef] [PubMed]
- Pirota, V.; Dell’Acqua, S.; Monzani, E.; Nicolis, S.; Casella, L. Copper-Aβ Peptides and Oxidation of Catecholic Substrates: Reactivity and Endogenous Peptide Damage. Chemistry 2016, 22, 16964–16973. [Google Scholar] [CrossRef]
- Dell’Acqua, S.; Pirota, V.; Monzani, E.; Camponeschi, F.; De Ricco, R.; Valensin, D.; Casella, L. Copper(I) Forms a Redox-Stable 1:2 Complex with α-Synuclein N-Terminal Peptide in a Membrane-Like Environment. Inorg. Chem. 2016, 55, 6100–6106. [Google Scholar] [CrossRef]
- Dell’Acqua, S.; Bacchella, C.; Monzani, E.; Nicolis, S.; Di Natale, G.; Rizzarelli, E.; Casella, L. Prion Peptides Are Extremely Sensitive to Copper Induced Oxidative Stress. Inorg. Chem. 2017, 56, 11317–11325. [Google Scholar] [CrossRef]
- Li, Y.; Jongberg, S.; Andersen, M.L.; Davies, M.J.; Lund, M.N. Quinone-induced protein modifications: Kinetic preference for reaction of 1,2-benzoquinones with thiol groups in proteins. Free Radic. Biol. Med. 2016, 97, 148–157. [Google Scholar] [CrossRef]
- Mitra, A.; Sarkar, N. The role of intra and inter-molecular disulfide bonds in modulating amyloidogenesis: A review. Arch. Biochem. Biophys. 2022, 716, 109113. [Google Scholar] [CrossRef]
- Herrera, A.; Muñoz, P.; Steinbusch, H.W.M.; Segura-Aguilar, J. Are Dopamine Oxidation Metabolites Involved in the Loss of Dopaminergic Neurons in the Nigrostriatal System in Parkinson’s Disease? ACS Chem. Neurosci. 2017, 8, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Bacchella, C.; Nicolis, S.; Dell’Acqua, S.; Rizzarelli, E.; Monzani, E.; Casella, L. Membrane Binding Strongly Affecting the Dopamine Reactivity Induced by Copper Prion and Copper/Amyloid-beta (Abeta) Peptides. A Ternary Copper/Abeta/Prion Peptide Complex Stabilized and Solubilized in Sodium Dodecyl Sulfate Micelles. Inorg. Chem. 2020, 59, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Griffiths, P.T.; Campbell, S.J.; Utinger, B.; Kalberer, M.; Paulson, S.E. Ascorbate oxidation by iron, copper and reactive oxygen species: Review, model development, and derivation of key rate constants. Sci. Rep. 2021, 11, 7417. [Google Scholar] [CrossRef]
- De Ricco, R.; Valensin, D.; Dell’Acqua, S.; Casella, L.; Hureau, C.; Faller, P. Copper(I/II), alpha/beta-Synuclein and Amyloid-beta: Menage a Trois? Chembiochem 2015, 16, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- Golec, C.; Mortensen, S.; Anwar, S.; Martic-Milne, S. Dual roles of tau R peptides on Cu(II)/(I)-mediated reactive oxygen species formation. J. Biol Inorg Chem 2021, 26, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Quaretti, M.; Porchia, M.; Tisato, F.; Trapananti, A.; Aquilanti, G.; Damjanovic, M.; Marchio, L.; Giorgetti, M.; Tegoni, M. Thermodynamic stability and structure in aqueous solution of the [Cu(PTA)4](+) complex (PTA=aminophosphine1,3,5triaza7phosphaadamantane). J. Inorg. Biochem. 2018, 188, 50–61. [Google Scholar] [CrossRef]
- Gran, G. Determination of the equivalence point in potentiometric titrations. Part II. The Analyst 1952, 77, 661–670. [Google Scholar] [CrossRef]
- Gans, P.; Sabatini, A.; Vacca, A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta 1996, 43, 1739–1753. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Góngora-Benítez, M.; Tulla-Puche, J.; Paradís-Bas, M.; Werbitzky, O.; Giraud, M.; Albericio, F. Optimized Fmoc solid-phase synthesis of the cysteine-rich peptide linaclotide. Biopolymers 2011, 96, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, C.W.; Hooker, J.M. Beyond the Amyloid Hypothesis of Alzheimer’s Disease: Tau Pathology Takes Center Stage. ACS Chem. Neurosci. 2018, 9, 2519. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K. Extracellular Tau and Its Potential Role in the Propagation of Tau Pathology. Front. Neurosci. 2017, 11, 667. [Google Scholar] [CrossRef]



| pKa (H4L4+) | Species | log β | ||
|---|---|---|---|---|
| pKa1 | 5.67(1) | NimH+ His | [CuH2L]4+ | 25.79(1) |
| pKa2 | 6.56(1) | NimH+ His | [CuHL]3+ | 19.42(2) |
| pKa3 | 9.83(1) | NH3+ Lys | [CuL]2+ | 12.68(2) |
| pKa4 | 10.50(1) | NH3+ Lys | [CuLH−1]+ | 4.09(3) |
| [CuLH−2] | −5.84(3) | |||
| [CuLH−3]− | −16.40(4) | |||
| Time | R3A | Oxidation | Dopamination | Oxidation and Dopamination | |||
|---|---|---|---|---|---|---|---|
| +16 | +32 | +149 | +151 | +149+16 | +151+16 | ||
| 10′ | 97 | 2 | - | 1 | - | - | - |
| 30′ | 92 | 5 | 1 | 1 | - | 1 | - |
| 60′ | 83 | 14 | 3 | 1 | - | - | - |
| 90′ | 63 | 27 | 6 | 1 | - | - | - |
| Time | R3C | Oxidation | Dopamination | Oxidation and Dopamination | Dimerization | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| +16 | +32 | +48 | +149 | +151 | +149 +149 | +151 +151 | +149 +16 | +151 +16 | |||
| 5′ | 29 | 1 | 1 | 1 | 9 | 52 | 1 | 1 | 1 | 2 | 2 |
| 10′ | 5 | 1 | 1 | 1 | 13 | 65 | 2 | 1 | 4 | 4 | 4 |
| 30′ | 2 | - | 1 | - | 12 | 70 | 2 | 2 | 3 | 3 | 4 |
| Time | R3C | Oxidation | Dopamination | Oxidation and Dopamination | Dimerization | -AcCys Addition | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| +16 | +32 | +48 | +149 | +151 | +149 +149 | +151 +151 | +149 +16 | +151 +16 | ||||
| 10′ | 42 | 1 | 1 | 1 | 5 | 34 | 1 | 1 | 2 | 1 | 2 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bacchella, C.; Gentili, S.; Mozzi, S.I.; Monzani, E.; Casella, L.; Tegoni, M.; Dell’Acqua, S. Role of the Cysteine in R3 Tau Peptide in Copper Binding and Reactivity. Int. J. Mol. Sci. 2022, 23, 10726. https://doi.org/10.3390/ijms231810726
Bacchella C, Gentili S, Mozzi SI, Monzani E, Casella L, Tegoni M, Dell’Acqua S. Role of the Cysteine in R3 Tau Peptide in Copper Binding and Reactivity. International Journal of Molecular Sciences. 2022; 23(18):10726. https://doi.org/10.3390/ijms231810726
Chicago/Turabian StyleBacchella, Chiara, Silvia Gentili, Sara Ida Mozzi, Enrico Monzani, Luigi Casella, Matteo Tegoni, and Simone Dell’Acqua. 2022. "Role of the Cysteine in R3 Tau Peptide in Copper Binding and Reactivity" International Journal of Molecular Sciences 23, no. 18: 10726. https://doi.org/10.3390/ijms231810726






