Cellular and Molecular Variations in Male and Female Murine Skeletal Muscle after Long-Term Feeding with a High-Fat Diet
Abstract
:1. Introduction
2. Results
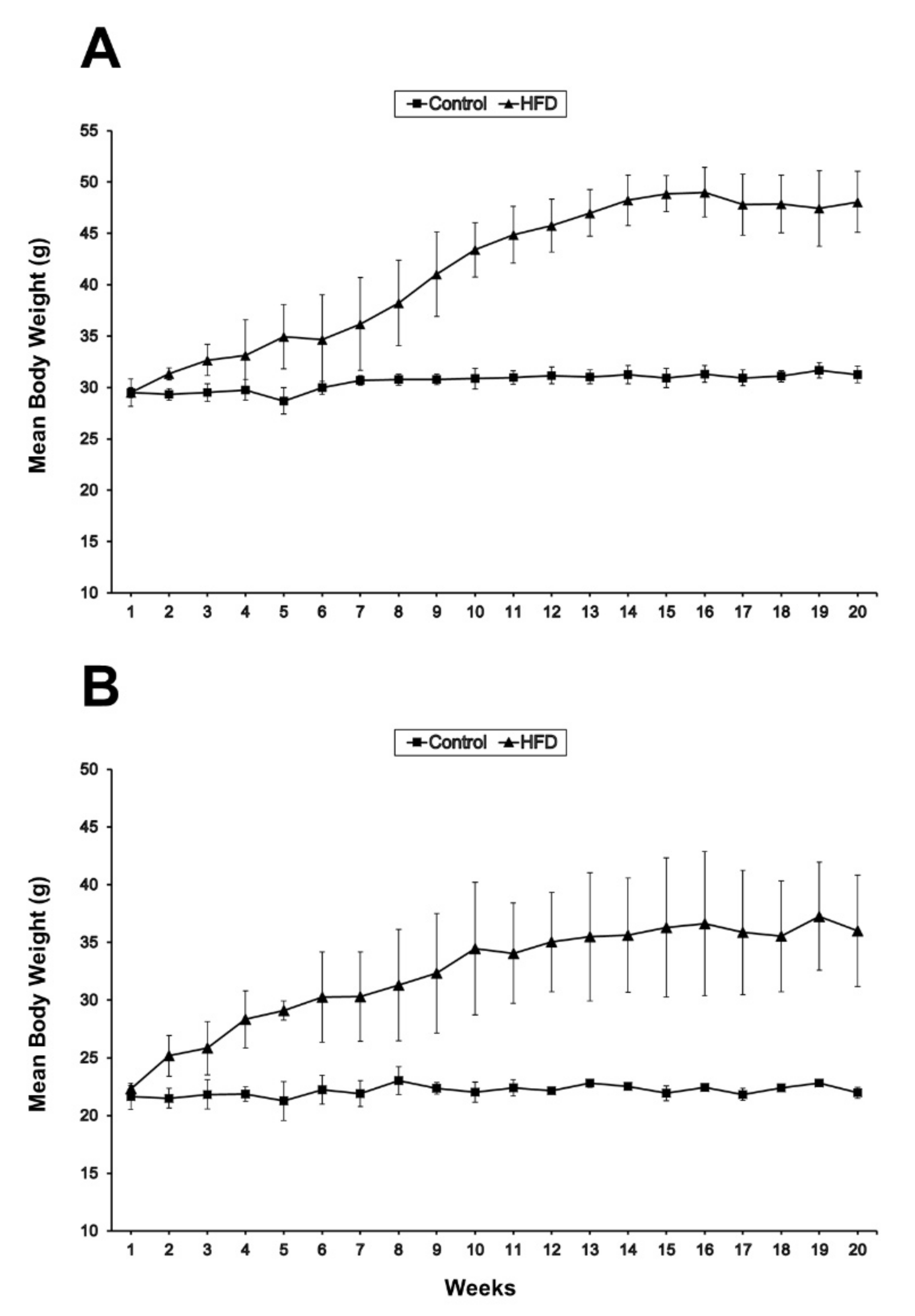
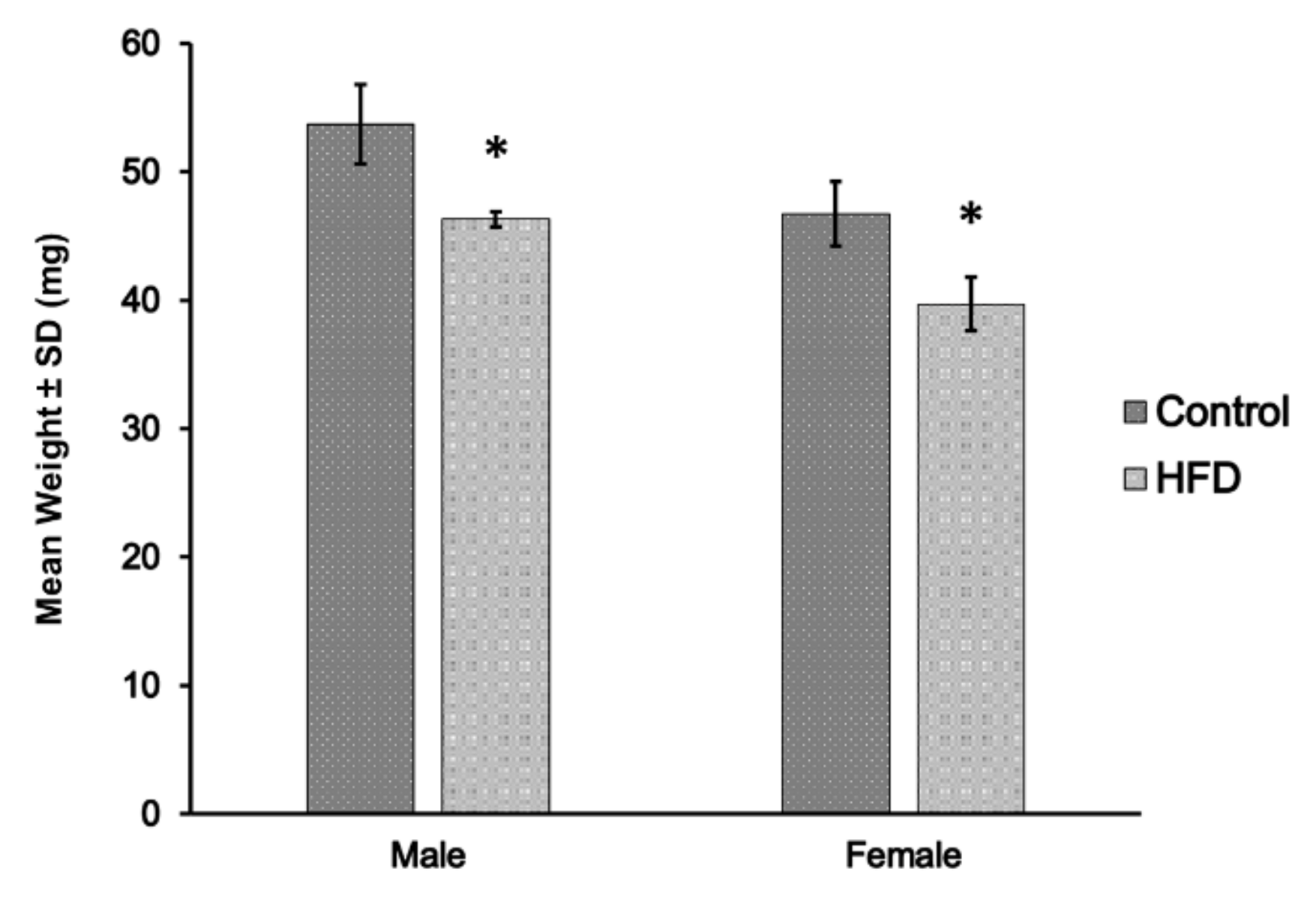
2.1. Body and Muscle Weights
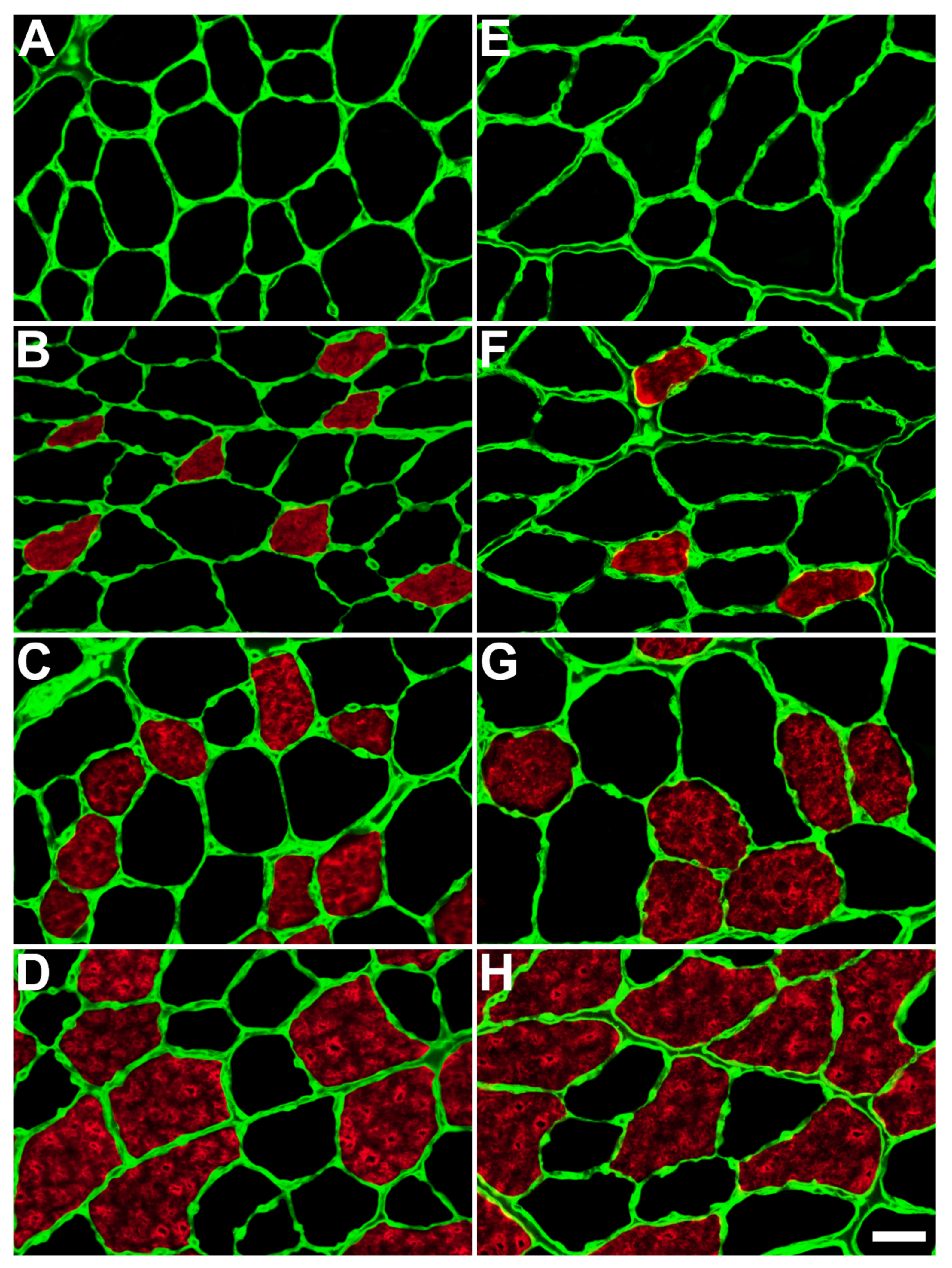
2.2. Fiber Morphology (Type, Size, and Intracellular Lipids)
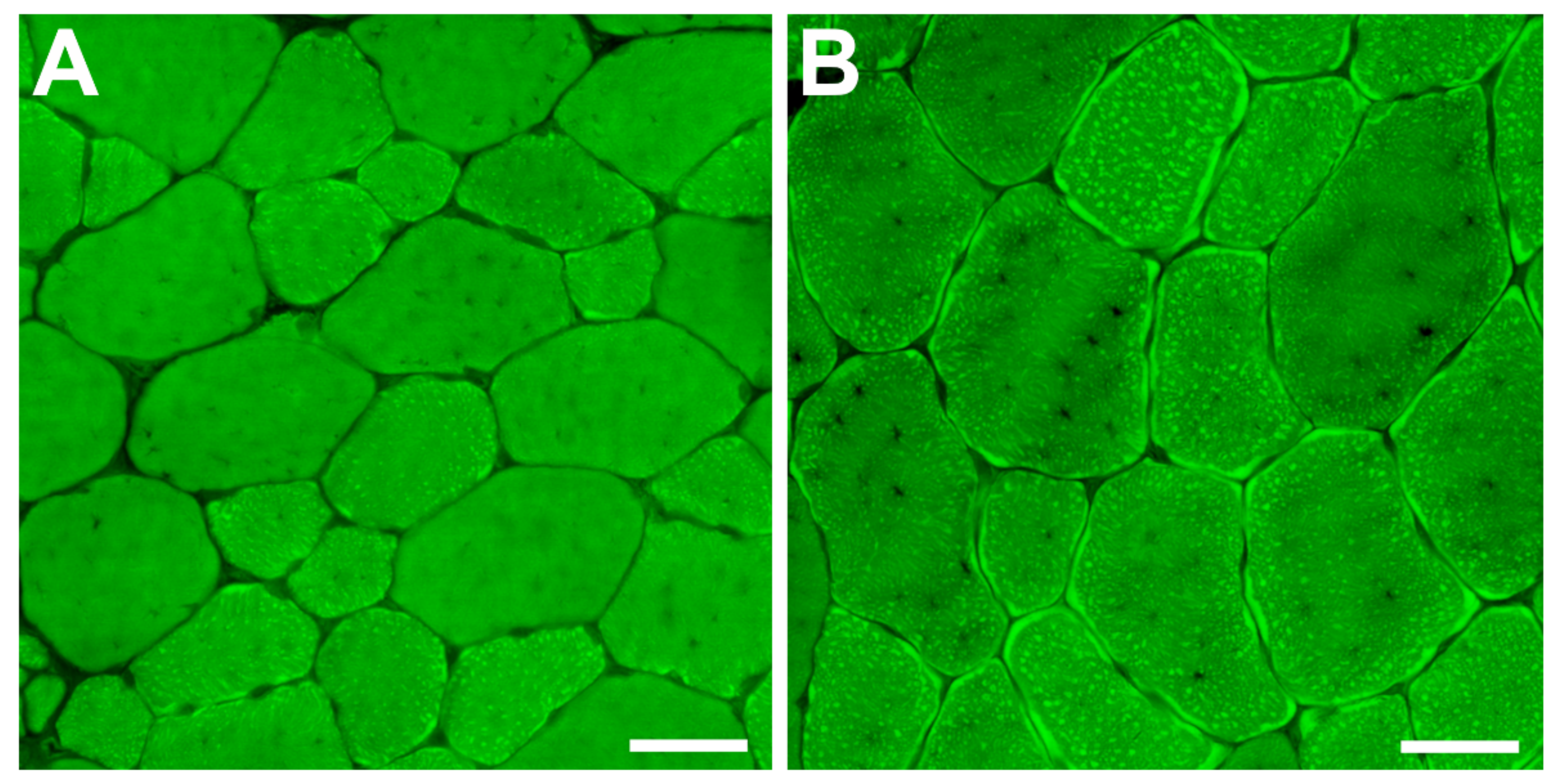
2.3. Satellite Cells and Myonuclei
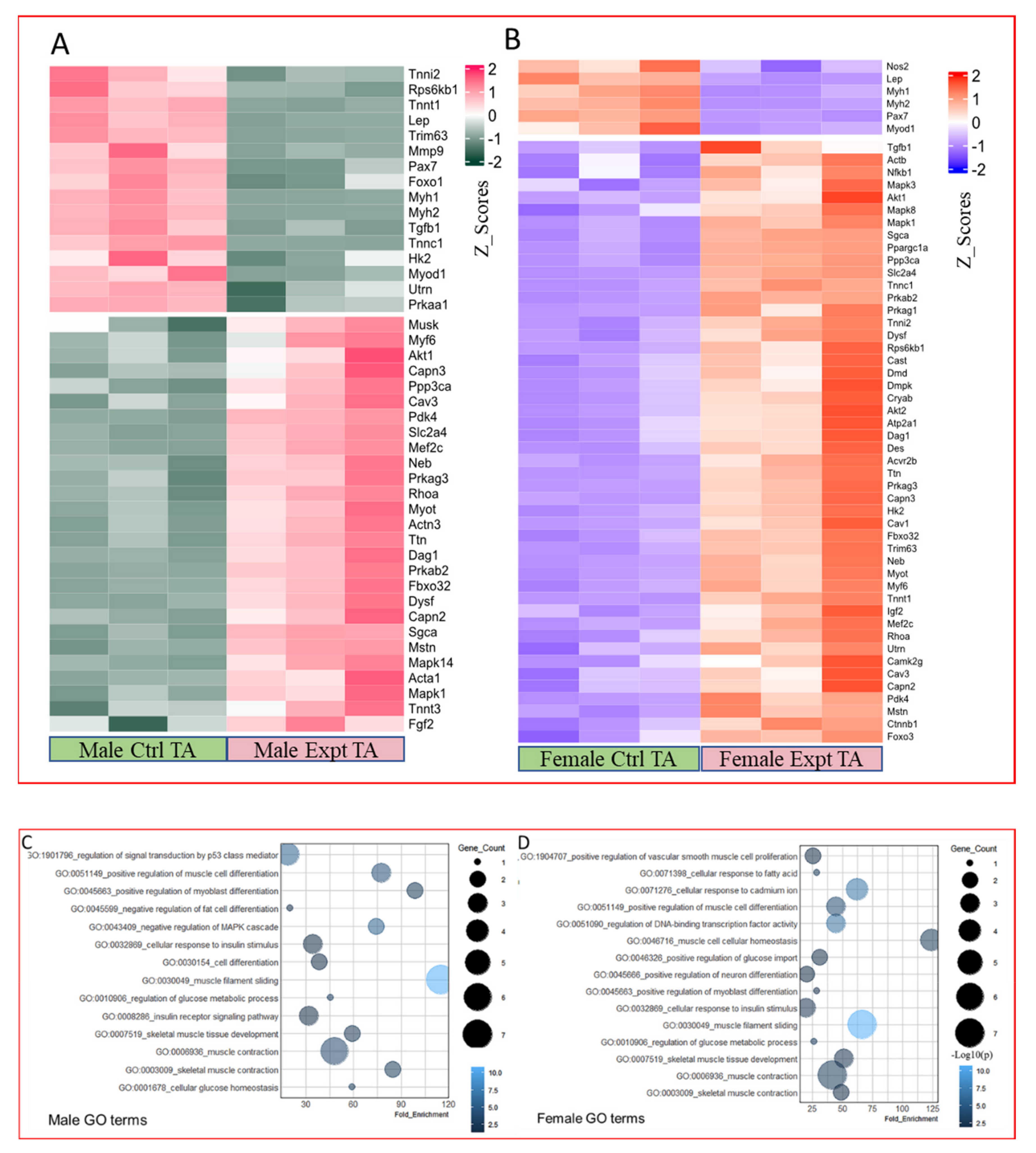
2.4. PCR Array
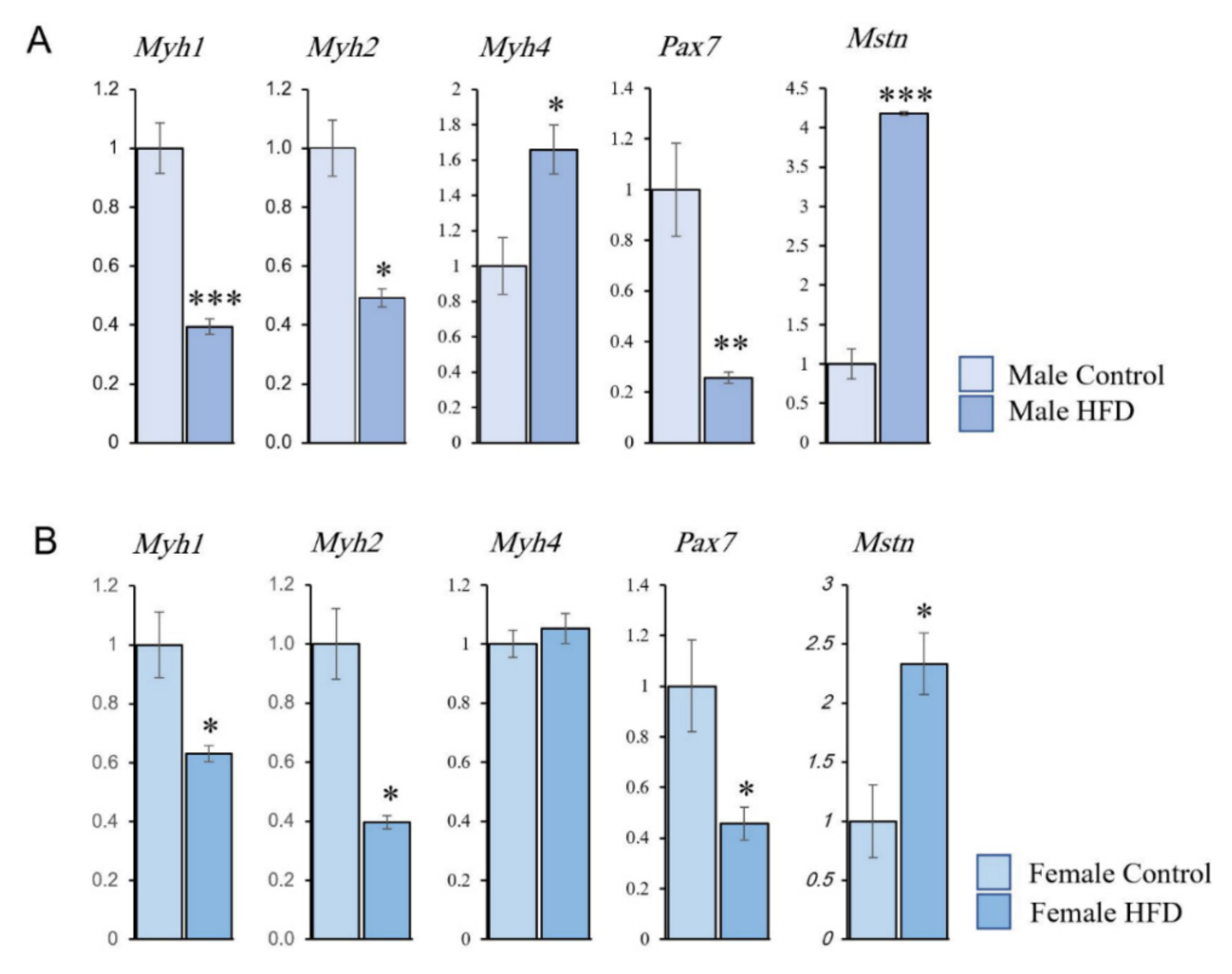
2.5. Quantitative Real-Time PCR
3. Discussion
4. Materials and Methods
4.1. Experimental Model
4.2. Tissue Preparation and Sectioning
4.3. Fiber Typing Analysis
4.3.1. Primary Antibodies
4.3.2. Immunocytochemical Protocol
4.3.3. Image Analysis and Data Collection
4.4. Labeling of Intracellular Lipids
4.4.1. Labeling Agent
4.4.2. Staining Protocol
4.5. Satellite Cell and Myonuclear Distribution
4.5.1. Primary Antibodies and Nuclear Labeling Agent
4.5.2. Immunocytochemical Protocol
4.5.3. Image Analysis and Data Collection
4.6. PCR Profiler Array for Myogenesis
4.6.1. RNA Preparation and Reverse Transcription
4.6.2. PCR Array
4.6.3. Bioinformatics Analysis
4.7. Real-Time Quantitative PCR
| Pax7 | |
| Forward primer | CAGCAAGCCCAGACAGGTG |
| Reverse primer | CCGGATTTCCCAGCTGAACA |
| Myh1 | |
| Forward primer | CACGCTGGATGCTGAGATTA |
| Reverse primer | AGGTGCAGCTGAGTGTCCTT |
| Myh2 | |
| Forward primer | GAATGCCTACGAGGAGTCTCT |
| Reverse primer | TTCTGCAATCTGTTCCGTGA |
| Myh4 | |
| Forward primer | CAAGTCATCGGTGTTTGTGG |
| Reverse primer | GGCCATGTCCTCAATCTTGT |
| Mstn | |
| Forward primer | CCCCCTCACGGTCGATTTTG |
| Reverse primer | GGTGCACAAGATGAGTATGCG |
| Gapdh | |
| Forward primer | GGTGCTGAGTATGTCGTGGAG |
| Reverse primer | TTCTCGTGGTTCACACCCAT |
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.Y.; Guo, C.C.; Yu, Y.X.; Xie, L.; Chang, C.Q. Establishment of high-fat diet-induced obesity and insulin resistance model in rats. J. Peking Univ. 2020, 52, 557–563. [Google Scholar] [CrossRef]
- Li, J.; Wu, H.; Liu, Y.; Yang, L. High Fat Diet Induced Obesity Model Using Four Strainsof Mice: Kunming, C57BL/6, BALB/c and ICR. Exp. Anim. 2020, 69, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Allouh, M.Z.; Jarrar, A.A.; Asfour, H.A.; Said, R.S.; Shaqoura, E.I. Sustanon Induces Dose-Independent Hypertrophy and Satellite Cell Proliferation in Slow Oxidative Fibers of Avian Skeletal Muscle. Histol. Histopathol. 2017, 32, 1151–1159. [Google Scholar] [CrossRef] [PubMed]
- Allouh, M.Z.; Yablonka-Reuveni, Z.; Rosser, B.W.C. Pax7 Reveals a Greater Frequency and Concentration of Satellite Cells at the Ends of Growing Skeletal Muscle Fibers. J. Histochem. Cytochem. 2008, 56, 77–87. [Google Scholar] [CrossRef]
- Allouh, M.Z.; Rosser, B.W.C. Nandrolone Decanoate Increases Satellite Cell Numbers in the Chicken Pectoralis Muscle. Histol. Histopathol. 2010, 25, 133–140. [Google Scholar] [CrossRef]
- Allouh, M.Z.; Aldirawi, M.H. Influence of Mesterolone on Satellite Cell Distribution and Fiber Morphology within Maturing Chicken Pectoralis Muscle. Anat. Rec. 2012, 295, 792–799. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Westerblad, H.; Bruton, J.D.; Katz, A. Skeletal Muscle: Energy Metabolism, Fiber Types, Fatigue and Adaptability. Exp. Cell Res. 2010, 316, 3093–3099. [Google Scholar] [CrossRef]
- Corcoran, M.P.; Lamon-Fava, S.; Fielding, R.A. Skeletal Muscle Lipid Deposition and Insulin Resistance: Effect of Dietary Fatty Acids and Exercise. Am. J. Clin. Nutr. 2007, 85, 662–677. [Google Scholar] [CrossRef]
- Shortreed, K.E.; Krause, M.P.; Huang, J.H.; Dhanani, D.; Moradi, J.; Ceddia, R.B.; Hawke, T.J. Muscle-Specific Adaptations, Impaired Oxidative Capacity and Maintenance of Contractile Function Characterize Diet-Induced Obese Mouse Skeletal Muscle. PLoS ONE 2009, 4, e7293. [Google Scholar] [CrossRef]
- Sparks, L.M.; Xie, H.; Koza, R.A.; Mynatt, R.; Hulver, M.W.; Bray, G.A.; Smith, S.R. A High-Fat Diet Coordinately Downregulates Genes Required for Mitochondrial Oxidative Phosphorylation in Skeletal Muscle. Diabetes 2005, 54, 1926–1933. [Google Scholar] [CrossRef]
- Choi, S.J.; Files, D.C.; Zhang, T.; Wang, Z.-M.; Messi, M.L.; Gregory, H.; Stone, J.; Lyles, M.F.; Dhar, S.; Marsh, A.P.; et al. Intramyocellular Lipid and Impaired Myofiber Contraction in Normal Weight and Obese Older Adults. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 557–564. [Google Scholar] [CrossRef]
- Lee, S.-R.; Khamoui, A.V.; Jo, E.; Park, B.-S.; Zourdos, M.C.; Panton, L.B.; Ormsbee, M.J.; Kim, J.-S. Effects of Chronic High-Fat Feeding on Skeletal Muscle Mass and Function in Middle-Aged Mice. Aging Clin. Exp. Res. 2015, 27, 403–411. [Google Scholar] [CrossRef]
- Thomas, M.M.; Trajcevski, K.E.; Coleman, S.K.; Jiang, M.; Di Michele, J.; O’Neill, H.M.; Lally, J.S.; Steinberg, G.R.; Hawke, T.J. Early Oxidative Shifts in Mouse Skeletal Muscle Morphology with High-Fat Diet Consumption Do Not Lead to Functional Improvements. Physiol. Rep. 2014, 2, e12149. [Google Scholar] [CrossRef]
- Hua, N.; Takahashi, H.; Yee, G.M.; Kitajima, Y.; Katagiri, S.; Kojima, M.; Anzai, K.; Eguchi, Y.; Hamilton, J.A. Influence of Muscle Fiber Type Composition on Early Fat Accumulation under High-Fat Diet Challenge. PLoS ONE 2017, 12, e0182430. [Google Scholar] [CrossRef]
- Hickey, M.S.; Carey, J.O.; Azevedo, J.L.; Houmard, J.A.; Pories, W.J.; Israel, R.G.; Dohm, G.L. Skeletal Muscle Fiber Composition is Related to Adiposity and In Vitro Glucose Transport Rate in Humans. Am. J. Physiol. 1995, 268, E453–E457. [Google Scholar] [CrossRef]
- Kriketos, A.D.; Pan, D.A.; Lillioja, S.; Cooney, G.J.; Baur, L.A.; Milner, M.R.; Sutton, J.R.; Jenkins, A.B.; Bogardus, C.; Storlien, L.H. Interrelationships between Muscle Morphology, Insulin Action, and Adiposity. Am. J. Physiol. 1996, 270, R1332–R1339. [Google Scholar] [CrossRef]
- Umek, N.; Horvat, S.; Cvetko, E. Skeletal Muscle and Fiber Type-Specific Intramyocellular Lipid Accumulation in Obese Mice. Bosn. J. Basic Med. Sci. 2021, 21, 730–738. [Google Scholar] [CrossRef]
- Asfour, H.A.; Shaqoura, E.I.; Said, R.S.; Mustafa, A.G.; Emerald, B.S.; Allouh, M.Z. Differential Response of Oxidative and Glycolytic Skeletal Muscle Fibers to Mesterolone. Sci. Rep. 2021, 11, 12301. [Google Scholar] [CrossRef]
- Fu, X.; Zhu, M.; Zhang, S.; Foretz, M.; Viollet, B.; Du, M. Obesity Impairs Skeletal Muscle Regeneration through Inhibition of AMPK. Diabetes 2016, 65, 188–200. [Google Scholar] [CrossRef]
- Woo, M.; Isganaitis, E.; Cerletti, M.; Fitzpatrick, C.; Wagers, A.J.; Jimenez-Chillaron, J.; Patti, M.E. Early Life Nutrition Modulates Muscle Stem Cell Number: Implications for Muscle Mass and Repair. Stem Cells Dev. 2011, 20, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, D.M.; Trajcevski, K.E.; Al-Sajee, D.; Wang, D.C.; Thomas, M.; Anderson, J.E.; Hawke, T.J. Diet-Induced Obesity Impairs Muscle Satellite Cell Activation and Muscle Repair through Alterations in Hepatocyte Growth Factor Signaling. Physiol. Rep. 2015, 3, e12506. [Google Scholar] [CrossRef] [PubMed]
- Asfour, H.A.; Allouh, M.Z.; Said, R.S. Myogenic Regulatory Factors: The Orchestrators of Myogenesis after 30 Years of Discovery. Exp. Biol. Med. 2018, 243, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Wang, H.; Lee, I.H.; Modi, S.; Wang, X.; Du, J.; Mitch, W.E. PTEN Inhibition Improves Muscle Regeneration in Mice Fed a High-Fat Diet. Diabetes 2010, 59, 1312–1320. [Google Scholar] [CrossRef]
- Akhmedov, D.; Berdeaux, R. The Effects of Obesity on Skeletal Muscle Regeneration. Front. Physiol. 2013, 4, 371. [Google Scholar] [CrossRef]
- Xu, P.; Werner, J.-U.; Milerski, S.; Hamp, C.M.; Kuzenko, T.; Jähnert, M.; Gottmann, P.; de Roy, L.; Warnecke, D.; Abaei, A.; et al. Diet-Induced Obesity Affects Muscle Regeneration after Murine Blunt Muscle Trauma—A Broad Spectrum Analysis. Front. Physiol. 2018, 9, 674. [Google Scholar] [CrossRef]
- Geiger, A.E.; Daughtry, M.R.; Yen, C.-N.; Kirkpatrick, L.T.; Shi, H.; Gerrard, D.E. Dual Effects of Obesity on Satellite Cells and Muscle Regeneration. Physiol. Rep. 2020, 8, e14511. [Google Scholar] [CrossRef]
- Sharma, M.; Langley, B.; Bass, J.; Kambadur, R. Myostatin in Muscle Growth and Repair. Exerc. Sport Sci. Rev. 2001, 29, 155–158. [Google Scholar] [CrossRef]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a Negative Regulator of Muscle Growth, Functions by Inhibiting Myoblast Proliferation. J. Biol. Chem. 2000, 275, 40235–40243. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.J. Regulation of Skeletal Muscle Mass in Mice by a New TGF-Beta Superfamily Member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.-J. Suppression of Body Fat Accumulation in Myostatin-Deficient Mice. J. Clin. Investig. 2002, 109, 595–601. [Google Scholar] [CrossRef]
- Schiaffino, S.; Rossi, A.C.; Smerdu, V.; Leinwand, L.A.; Reggiani, C. Developmental Myosins: Expression Patterns and Functional Significance. Skelet. Muscle 2015, 5, 22. [Google Scholar] [CrossRef]
- Medler, S. Mixing it up: The Biological Significance of Hybrid Skeletal Muscle Fibers. J. Exp. Biol. 2019, 222, jeb200832. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, Y.; Sun, Q.; Yu, L.; Li, M.; Zheng, B.; Wu, X.; Yang, B.; Li, Y.; Huang, C. Extract of Okra Lowers Blood Glucose and Serum Lipids in High-Fat Diet-Induced Obese C57BL/6 Mice. J. Nutr. Biochem. 2014, 25, 702–709. [Google Scholar] [CrossRef]
- Mong, M.; Chao, C.; Yin, M. Histidine and Carnosine Alleviated Hepatic Steatosis in Mice Consumed High Saturated Fat Diet. Eur. J. Pharmacol. 2011, 653, 82–88. [Google Scholar] [CrossRef]
- Walewski, J.L.; Ge, F.; Lobdell, H.; Levin, N.; Schwartz, G.J.; Vasselli, J.R.; Pomp, A.; Dakin, G.; Berk, P.D. Spexin is a Novel Human Peptide that Reduces Adipocyte Uptake of Long Chain Fatty Acids and Causes Weight Loss in Rodents with Diet-Induced Obesity. Obesity 2014, 22, 1643–1652. [Google Scholar] [CrossRef]
- Tyagi, S.; Beqollari, D.; Lee, C.S.; Walker, L.A.; Bannister, R.A. Semi-Automated Analysis of Mouse Skeletal Muscle Morphology and Fiber-Type Composition. J. Vis. Exp. 2017, 126, e56024. [Google Scholar] [CrossRef]
- Allouh, M.Z. Effect of Ferula Hermonis Root Extract on Rat Skeletal Muscle Adaptation to Exercise. Exp. Biol. Med. 2011, 236, 1373–1378. [Google Scholar] [CrossRef]
- Spangenburg, E.E.; Pratt, S.J.P.; Wohlers, L.M.; Lovering, R.M. Use of BODIPY (493/503) to Visualize Intramuscular Lipid Droplets in Skeletal Muscle. J. Biomed. Biotechnol. 2011, 2011, 598358. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An Improvement of the 2^(-Delta Delta CT) Method for Quantitative Real-Time Polymerase Chain Reaction Data Analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Ulgen, E.; Ozisik, O.; Sezerman, O.U. PathfindR: An R Package for Comprehensive Identification of Enriched Pathways in Omics Data through Active Subnetworks. Front. Genet. 2019, 10, 858. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]


| Fiber Type | Fiber Frequency (%) | Fiber Diameter (μm) | ||
|---|---|---|---|---|
| Control | HFD | Control | HFD | |
| I | 0.0 | 0.0 | - | - |
| IIA | 10.9 ± 1.0 | 7.4 ± 2.0 * | 22.9 ± 1.9 | 30.8 ± 1.4 * |
| IIX | 37.8 ± 1.5 | 32.7 ± 2.3 * | 29.3 ± 1.0 | 35.5 ± 3.3 * |
| IIB | 50.2 ± 4.3 | 59.5 ± 5.9 | 41.8 ± 0.5 | 41.6 ± 1.0 |
| Fiber Type | Fiber Frequency (%) | Fiber Diameter (μm) | ||
|---|---|---|---|---|
| Control | HFD | Control | HFD | |
| I | 0.0 | 0.0 | - | - |
| IIA | 15.0 ± 1.3 | 8.6 ± 2.8 * | 24.7 ± 2.5 | 24.9 ± 3.0 |
| IIX | 40.7 ± 8.1 | 32.8 ± 7.6 | 27.2 ± 1.0 | 32.8 ± 2.6 * |
| IIB | 44.4 ± 6.4 | 57.8 ± 13.8 | 38.6 ± 3.2 | 39.0 ± 5.1 |
| Sex | Male | Female | ||
|---|---|---|---|---|
| Group | Control | HFD | Control | HFD |
| Fiber Diameter # (μm) | 36.3 ± 0.7 | 41.1 ± 2.5 * | 34.1 ± 1.7 | 38.0 ± 1.1 * |
| Length of SCN (μm) | 9.9 ± 0.1 | 9.5 ± 0.4 | 10.3 ± 0.2 | 9.9 ± 0.2 |
| SC Frequency (%) | 7.0 ± 0.2 | 3.7 ± 0.1 ** | 7.3 ± 0.2 | 3.6 ± 0.2 ** |
| SCs/mm | 4.9 ± 0.4 | 1.8 ± 0.1 ** | 4.7 ± 0.5 | 1.7 ± 0.2 ** |
| Surface area/SC (×103 μm2) | 23.6 ± 2.3 | 73.1 ± 4.5 ** | 23.1 ± 2.6 | 70.6 ± 6.5 ** |
| Length of MN (μm) | 10.4 ± 0.1 | 10.1 ± 0.5 | 10.1 ± 0.4 | 10.2 ± 0.5 |
| MN/mm | 63.6 ± 4.9 | 44.7 ± 1.1 ** | 61.4 ± 4.0 | 42.9 ± 3.8 ** |
| MD (×103 μm3) | 16.4 ± 1.9 | 29.8 ± 4.0 ** | 15.0 ± 2.0 | 26.5 ± 2.0 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emerald, B.S.; Al Jailani, M.A.; Ibrahim, M.F.; Kumar, C.A.; Allouh, M.Z. Cellular and Molecular Variations in Male and Female Murine Skeletal Muscle after Long-Term Feeding with a High-Fat Diet. Int. J. Mol. Sci. 2022, 23, 9547. https://doi.org/10.3390/ijms23179547
Emerald BS, Al Jailani MA, Ibrahim MF, Kumar CA, Allouh MZ. Cellular and Molecular Variations in Male and Female Murine Skeletal Muscle after Long-Term Feeding with a High-Fat Diet. International Journal of Molecular Sciences. 2022; 23(17):9547. https://doi.org/10.3390/ijms23179547
Chicago/Turabian StyleEmerald, Bright Starling, Mohammed A. Al Jailani, Marwa F. Ibrahim, Challagandla Anil Kumar, and Mohammed Z. Allouh. 2022. "Cellular and Molecular Variations in Male and Female Murine Skeletal Muscle after Long-Term Feeding with a High-Fat Diet" International Journal of Molecular Sciences 23, no. 17: 9547. https://doi.org/10.3390/ijms23179547






