miRNA Expression Is Increased in Serum from Patients with Semantic Variant Primary Progressive Aphasia
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Population
4.2. Serum Sample Collection
4.3. AD CSF Biomarkers Measurement
4.4. miRNA Isolation from Human Serum Samples
4.5. Screening of miRNAs in Human Serum by Human Serum & Plasma miScript miRNA PCR Array
4.6. Validation of Best Hits by Taqman qRT-PCR Assays
4.7. Target Gene Prediction and Bioinformatics Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gorno-Tempini, M.L.; Hillis, A.E.; Weintraub, S.; Kertesz, A.; Mendez, M.; Cappa, S.F.; Ogar, J.M.; Rohrer, J.D.; Black, S.; Boeve, B.F.; et al. Classification of primary progressive aphasia and its variants. Neurology 2011, 76, 1006–1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tee, B.L.; Gorno-Tempini, M.L. Primary progressive aphasia: A model for neurodegenerative disease. Curr. Opin. Neurol. 2019, 32, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, E.G.; Mandelli, M.L.; Miller, Z.A.; Santos-Santos, M.A.; Wilson, S.M.; Agosta, F.; Grinberg, L.T.; Huang, E.J.; Trojanowski, J.Q.; Meyer, M.; et al. Typical and atypical pathology in primary progressive aphasia variants. Ann. Neurol. 2017, 81, 430–443. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, I.R.A.; Neumann, M.; Baborie, A.; Sampathu, D.M.; Du Plessis, D.; Jaros, E.; Perry, R.H.; Trojanowski, J.Q.; Mann, D.M.A.; Lee, V.M.Y. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011, 122, 111–113. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.B.; Porta, S.; Michael Baer, G.; Xu, Y.; Suh, E.R.; Kwong, L.K.; Elman, L.; Grossman, M.; Lee, V.M.Y.; Irwin, D.J.; et al. Expansion of the classification of FTLD-TDP: Distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol. 2017, 134, 65–78. [Google Scholar] [CrossRef]
- Ruksenaite, J.; Volkmer, A.; Jiang, J.; Johnson, J.C.; Marshall, C.R.; Warren, J.D.; Hardy, C.J. Primary Progressive Aphasia: Toward a Pathophysiological Synthesis. Curr. Neurol. Neurosci. Rep. 2021, 21, 7. [Google Scholar] [CrossRef]
- Mesulam, M.M.; Lalehzari, N.; Rahmani, F.; Ohm, D.; Shahidehpour, R.; Kim, G.; Gefen, T.; Weintraub, S.; Bigio, E.; Geula, C. Cortical cholinergic denervation in primary progressive aphasia with Alzheimer pathology. Neurology 2019, 92, E1580–E1588. [Google Scholar] [CrossRef]
- Nguyen, T.P.N.; Kumar, M.; Fedele, E.; Bonanno, G.; Bonifacino, T. MicroRNA Alteration, Application as Biomarkers, and Therapeutic Approaches in Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 4718. [Google Scholar] [CrossRef]
- Fenoglio, C.; Scarpini, E.; Serpente, M.; Galimberti, D. Role of Genetics and Epigenetics in the Pathogenesis of Alzheimer’s Disease and Frontotemporal Dementia. J. Alzheimer’s Dis. 2018, 62, 913–932. [Google Scholar] [CrossRef] [Green Version]
- Belzil, V.V.; Gendron, T.F.; Petrucelli, L. RNA-mediated toxicity in neurodegenerative disease. Mol. Cell. Neurosci. 2013, 56, 406–419. [Google Scholar] [CrossRef] [Green Version]
- Lau, P.; Bossers, K.; Janky, R.; Salta, E.; Frigerio, C.S.; Barbash, S.; Rothman, R.; Sierksma, A.S.R.; Thathiah, A.; Greenberg, D.; et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 1613–1634. [Google Scholar] [CrossRef] [Green Version]
- Galimberti, D.; Villa, C.; Fenoglio, C.; Serpente, M.; Ghezzi, L.; Cioffi, S.M.G.; Arighi, A.; Fumagalli, G.; Scarpini, E. Circulating miRNAs as Potential Biomarkers in Alzheimer’s Disease. J. Alzheimer’s Dis. 2014, 42, 1261–1267. [Google Scholar] [CrossRef]
- Fenoglio, C.; Ridolfi, E.; Cantoni, C.; De Riz, M.; Bonsi, R.; Serpente, M.; Villa, C.; Pietroboni, A.M.; Naismith, R.T.; Alvarez, E.; et al. Decreased circulating miRNA levels in patients with primary progressive multiple sclerosis. Mult. Scler. 2013, 19, 1938–1942. [Google Scholar] [CrossRef]
- Piscopo, P.; Grasso, M.; Puopolo, M.; D’Acunto, E.; Talarico, G.; Crestini, A.; Gasparini, M.; Campopiano, R.; Gambardella, S.; Castellano, A.E.; et al. Circulating miR-127-3p as a potential biomarker for differential diagnosis in frontotemporal dementia. J. Alzheimer’s Dis. 2018, 65, 455–464. [Google Scholar] [CrossRef]
- Grasso, M.; Piscopo, P.; Talarico, G.; Ricci, L.; Crestini, A.; Tosto, G.; Gasparini, M.; Bruno, G.; Denti, M.A.; Confaloni, A. Plasma microRNA profiling distinguishes patients with frontotemporal dementia from healthy subjects. Neurobiol. Aging 2019, 84, 240.e1–240.e12. [Google Scholar] [CrossRef]
- Kmetzsch, V.; Anquetil, V.; Saracino, D.; Rinaldi, D.; Camuzat, A.; Gareau, T.; Jornea, L.; Forlani, S.; Couratier, P.; Wallon, D.; et al. Plasma microRNA signature in presymptomatic and symptomatic subjects with C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2021, 92, 485–493. [Google Scholar] [CrossRef]
- Bergeron, D.; Gorno-Tempini, M.L.; Rabinovici, G.D.; Santos-Santos, M.A.; Seeley, W.; Miller, B.L.; Pijnenburg, Y.; Keulen, M.A.; Groot, C.; Van Berckel, B.N.M.; et al. Prevalence of amyloid-β pathology in distinct variants of primary progressive aphasia. Ann. Neurol. 2018, 84, 729–740. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.H.; Liu, Y.N. Downregulated serum miR-223 servers as biomarker in Alzheimer’s disease. Cell Biochem. Funct. 2016, 34, 233–237. [Google Scholar] [CrossRef]
- Giuliani, A.; Gaetani, S.; Sorgentoni, G.; Agarbati, S.; Laggetta, M.; Matacchione, G.; Gobbi, M.; Rossi, T.; Galeazzi, R.; Piccinini, G.; et al. Circulating Inflamma-miRs as Potential Biomarkers of Cognitive Impairment in Patients Affected by Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 647015. [Google Scholar] [CrossRef]
- Liu, L.; Liu, L.; Lu, Y.; Zhang, T.; Zhao, W. Serum aberrant expression of miR-24-3p and its diagnostic value in Alzheimer’s disease. Biomark. Med. 2021, 15, 1499–1507. [Google Scholar] [CrossRef]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma Exosomal miRNAs in Persons with and without Alzheimer Disease: Altered Expression and Prospects for Biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Xu, Y.; Wang, B.; Huang, J.; Li, Q. miR-34a-5p and miR-125b-5p attenuate Aβ-induced neurotoxicity through targeting BACE1. J. Neurol. Sci. 2020, 413, 116793. [Google Scholar] [CrossRef]
- Harris, J.M.; Gall, C.; Thompson, J.C.; Richardson, A.M.T.; Neary, D.; Du Plessis, D.; Pal, P.; Mann, D.M.A.; Snowden, J.S.; Jones, M. Classification and pathology of primary progressive aphasia. Neurology 2013, 81, 1832–1839. [Google Scholar] [CrossRef]
- De Boer, E.M.J.; Orie, V.K.; Williams, T.; Baker, M.R.; De Oliveira, H.M.; Polvikoski, T.; Silsby, M.; Menon, P.; Van Den Bos, M.; Halliday, G.M.; et al. TDP-43 proteinopathies: A new wave of neurodegenerative diseases. J. Neurol. Neurosurg. Psychiatry 2021, 92, 86–95. [Google Scholar] [CrossRef]
- Prasad, A.; Bharathi, V.; Sivalingam, V.; Girdhar, A.; Patel, B.K. Molecular mechanisms of TDP-43 misfolding and pathology in amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2019, 12, 25. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, J.; Cui, P.; Zhou, Y.; Liu, C.; Wu, X.; Ji, Y.; Wang, S.; Cheng, B.; Ye, H.; et al. TREM2 sustains macrophage-hepatocyte metabolic coordination in nonalcoholic fatty liver disease and sepsis. J. Clin. Investig. 2021, 131, e135197. [Google Scholar] [CrossRef]
- Prater, K.E.; Latimer, C.S.; Jayadev, S. Glial TDP-43 and TDP-43 induced glial pathology, focus on neurodegenerative proteinopathy syndromes. Glia 2022, 70, 239–255. [Google Scholar] [CrossRef]
- Bian, Y.; Pang, P.; Li, X.; Yu, S.; Wang, X.; Liu, K.; Ju, J.; Wu, H.; Gao, Y.; Liu, Q.; et al. CircHelz activates NLRP3 inflammasome to promote myocardial injury by sponging miR-133a-3p in mouse ischemic heart. J. Mol. Cell. Cardiol. 2021, 158, 128–139. [Google Scholar] [CrossRef]
- Starhof, C.; Hejl, A.M.; Heegaard, N.H.H.; Carlsen, A.L.; Burton, M.; Lilje, B.; Winge, K. The biomarker potential of cell-free microRNA from cerebrospinal fluid in Parkinsonian Syndromes. Mov. Disord. 2019, 34, 246–254. [Google Scholar] [CrossRef]
- Saǧlr, F.; Ersoy Tunall, N.; Tombul, T.; Koral, G.; Çlrak, S.; Yllmaz, V.; Türkoǧlu, R.; Tüzün, E.; Sağır, F.; Ersoy Tunalı, N.; et al. miR-132-3p, miR-106b-5p, and miR-19b-3p Are Associated with Brain-Derived Neurotrophic Factor Production and Clinical Activity in Multiple Sclerosis: A Pilot Study. Genet. Test. Mol. Biomark. 2021, 25, 720–726. [Google Scholar] [CrossRef]
- De Rossi, P.; Lewis, A.J.; Furrer, J.; De Vos, L.; Demeter, T.; Elie Zbinden, A.; Zhong, W.; Wiersma, V.I.; Scialo, C.; Weber, J.; et al. FTLD-TDP assemblies seed neoaggregates with subtype-specific features via a prion-like cascade. EMBO Rep. 2021, 22, e53877. [Google Scholar] [CrossRef] [PubMed]
- Raheja, R.; Regev, K.; Healy, B.C.; Mazzola, M.A.; Beynon, V.; Von Glehn, F.; Paul, A.; Diaz-Cruz, C.; Gholipour, T.; Glanz, B.I.; et al. Correlating serum micrornas and clinical parameters in amyotrophic lateral sclerosis. Muscle Nerve 2018, 58, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Mesulam, M.M.; Weintraub, S.; Rogalski, E.J.; Wieneke, C.; Geula, C.; Bigio, E.H. Asymmetry and heterogeneity of Alzheimer’s and frontotemporal pathology in primary progressive aphasia. Brain 2014, 137, 1176–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossman, M. Primary progressive aphasia: Clinicopathological correlations. Nat. Rev. Neurol. 2010, 6, 88–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.Y.; Ou, R.W.; Chen, Y.P.; Gu, X.J.; Wei, Q.Q.; Cao, B.; Zhang, L.Y.; Hou, Y.B.; Liu, K.C.; Chen, X.P.; et al. Mutation analysis of seven SLC family transporters for early-onset Parkinson’s disease in Chinese population. Neurobiol. Aging 2021, 103, 152.e1–152.e6. [Google Scholar] [CrossRef] [PubMed]
- Cibulka, M.; Brodnanova, M.; Grendar, M.; Necpal, J.; Benetin, J.; Han, V.; Kurca, E.; Nosal, V.; Skorvanek, M.; Vesely, B.; et al. Alzheimer’s Disease-Associated SNP rs708727 in SLC41A1 May Increase Risk for Parkinson’s Disease: Report from Enlarged Slovak Study. Int. J. Mol. Sci. 2022, 23, 1604. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.W.; Baker, M.C.; Jackson, J.L.; Dejesus-Hernandez, M.; Finch, N.C.A.; Tian, S.; Heckman, M.G.; Pottier, C.; Gendron, T.F.; Murray, M.E.; et al. Extensive transcriptomic study emphasizes importance of vesicular transport in C9orf72 expansion carriers. Acta Neuropathol. Commun. 2019, 7, 150. [Google Scholar] [CrossRef]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1?SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, T.; Murao, N.; Kadowaki, H.; Takao, K.; Miyakawa, T.; Matsushita, Y.; Katagiri, T.; Futatsugi, A.; Shinmyo, Y.; Kawasaki, H.; et al. ERAD components Derlin-1 and Derlin-2 are essential for postnatal brain development and motor function. iScience 2021, 24, 102758. [Google Scholar] [CrossRef]
- Vasudevan, S. Functional validation of microRNA-target RNA interactions. Methods 2012, 58, 126–134. [Google Scholar] [CrossRef]
- Sorrentino, F.; Arighi, A.; Serpente, M.; Arosio, B.; Arcaro, M.; Visconte, C.; Rotondo, E.; Vimercati, R.; Ferri, E.; Fumagalli, G.G.; et al. Niemann-Pick Type C 1 (NPC1) and NPC2 gene variability in demented patients with evidence of brain amyloid deposition. J. Alzheimer’s Dis. 2021, 83, 1313–1323. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
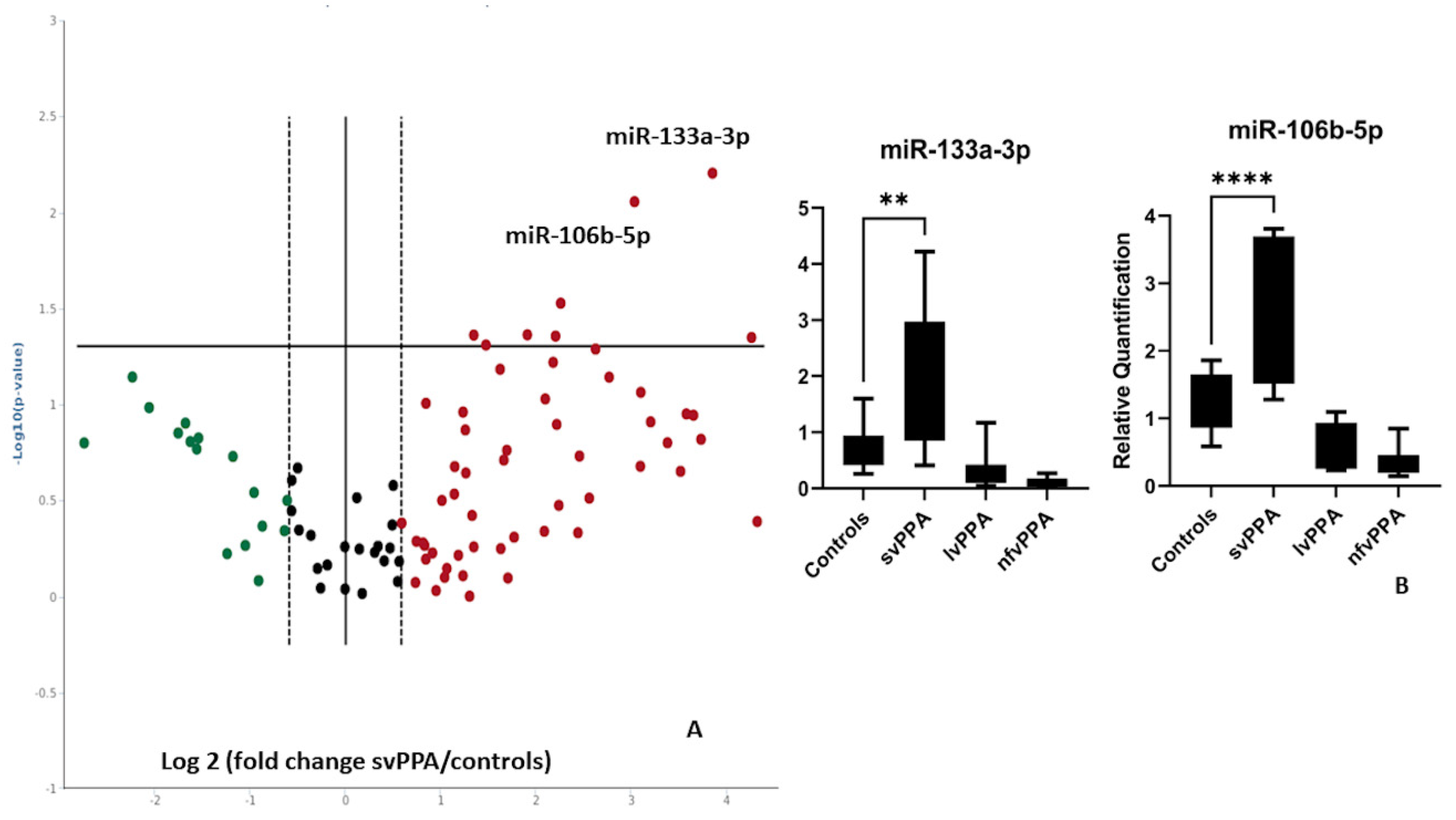

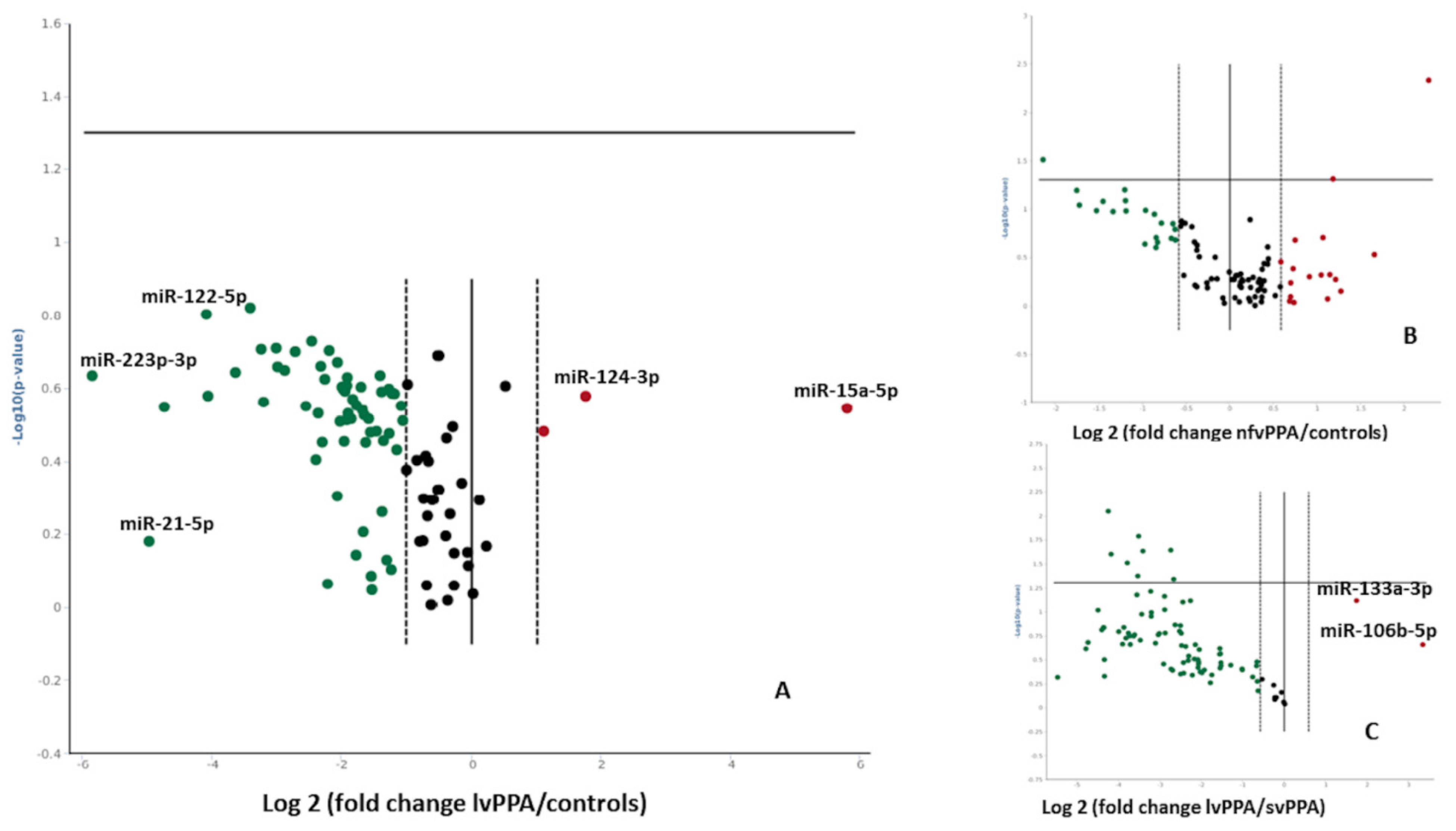
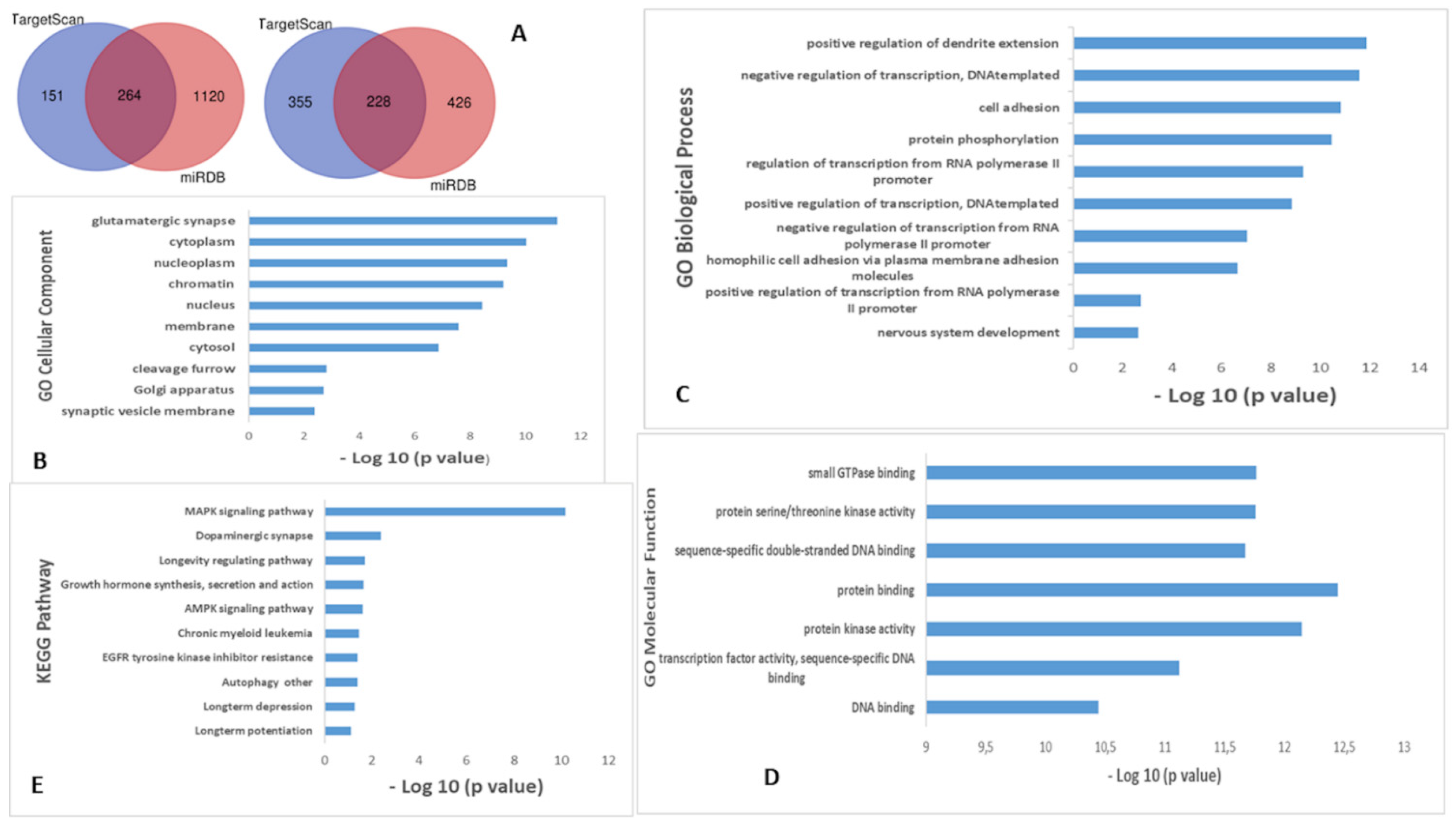
| Nonfluent/Agrammatic PPA (n = 18) | Logopenic PPA (n = 18) | Semantic Variant PPA (n = 18) | Healthy Controls (n = 18) | |
|---|---|---|---|---|
| gender (M:F) | 6:12 | 8:10 | 9:9 | 10:8 |
| Aβ42 (pg/mL; mean ± SD) | 723.6 ± 170.59 * | 571 ± 433.7 ** | 893.15 ± 325.6 *** | 1054.54 ± 252 |
| tau (pg/mL; mean ± SD) | 491.4 ± 177.50 | 456.4 ± 177.9 | 338.15 ± 170.2 | 380 ± 274 |
| p-tau (pg/mL; mean ± SD) | 55.6 ± 25.88 | 61.4 ± 24.27 | 49.38 ± 19.05 | 58 ± 46 |
| mean age (years ± SD) | 76.3 ± 8.6 | 81 ± 8.1 | 79 ± 9.4 | 78 ± 7.3 |
| mean age at onset (years ± SD) | 63.5 ± 0.44 | 70 ± 0.36 | 64 ± 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serpente, M.; Ghezzi, L.; Fenoglio, C.; Buccellato, F.R.; Fumagalli, G.G.; Rotondo, E.; Arcaro, M.; Arighi, A.; Galimberti, D. miRNA Expression Is Increased in Serum from Patients with Semantic Variant Primary Progressive Aphasia. Int. J. Mol. Sci. 2022, 23, 8487. https://doi.org/10.3390/ijms23158487
Serpente M, Ghezzi L, Fenoglio C, Buccellato FR, Fumagalli GG, Rotondo E, Arcaro M, Arighi A, Galimberti D. miRNA Expression Is Increased in Serum from Patients with Semantic Variant Primary Progressive Aphasia. International Journal of Molecular Sciences. 2022; 23(15):8487. https://doi.org/10.3390/ijms23158487
Chicago/Turabian StyleSerpente, Maria, Laura Ghezzi, Chiara Fenoglio, Francesca R. Buccellato, Giorgio G. Fumagalli, Emanuela Rotondo, Marina Arcaro, Andrea Arighi, and Daniela Galimberti. 2022. "miRNA Expression Is Increased in Serum from Patients with Semantic Variant Primary Progressive Aphasia" International Journal of Molecular Sciences 23, no. 15: 8487. https://doi.org/10.3390/ijms23158487






