Molecular Modelling of NONO and SFPQ Dimerization Process and RNA Recognition Mechanism
Abstract
:1. Introduction
2. Results and Discussion
2.1. Comparison of NONO and SFPQ Dimer Stability
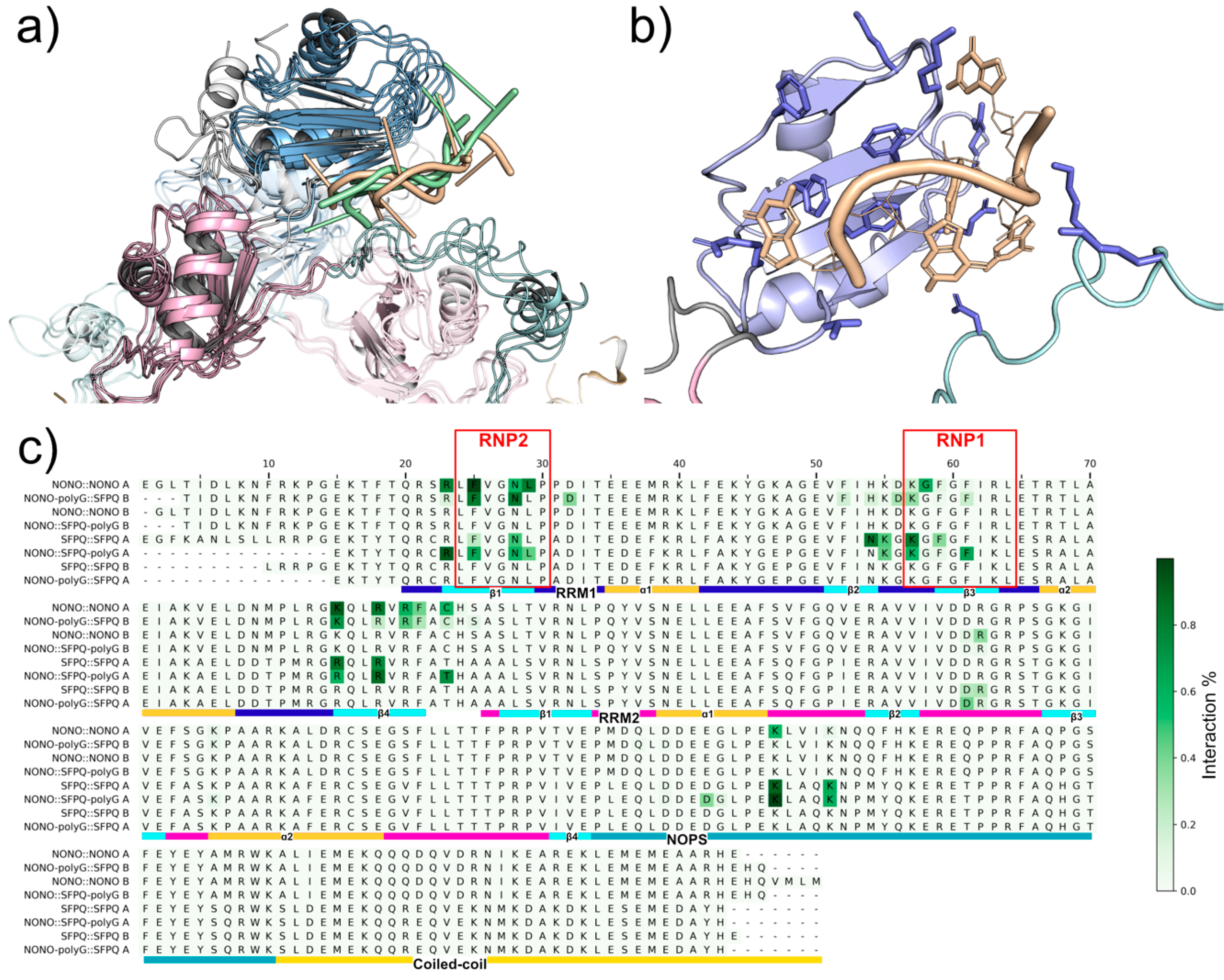
2.2. NONO and SFPQ Dimers Have Different Interaction Patterns
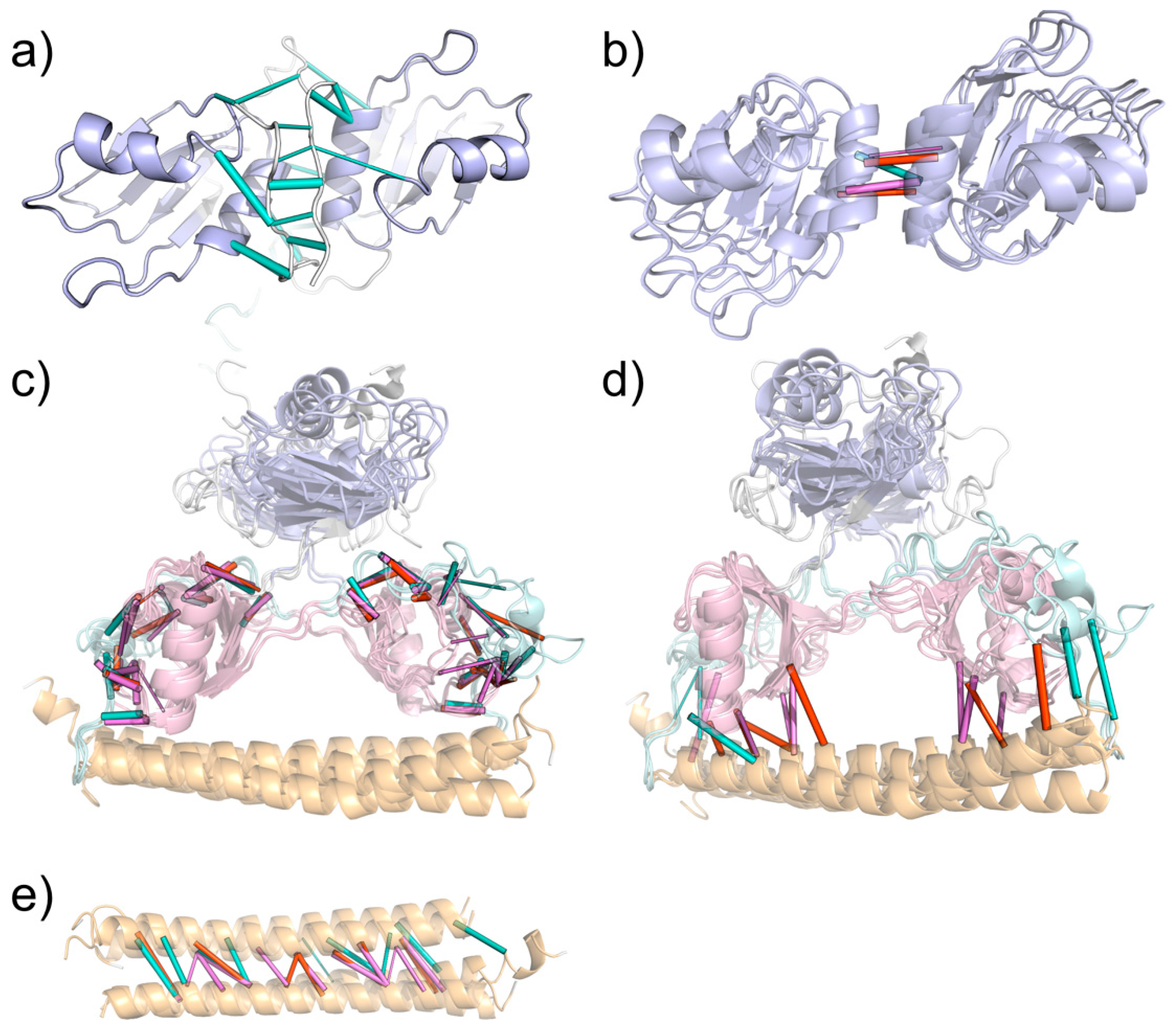
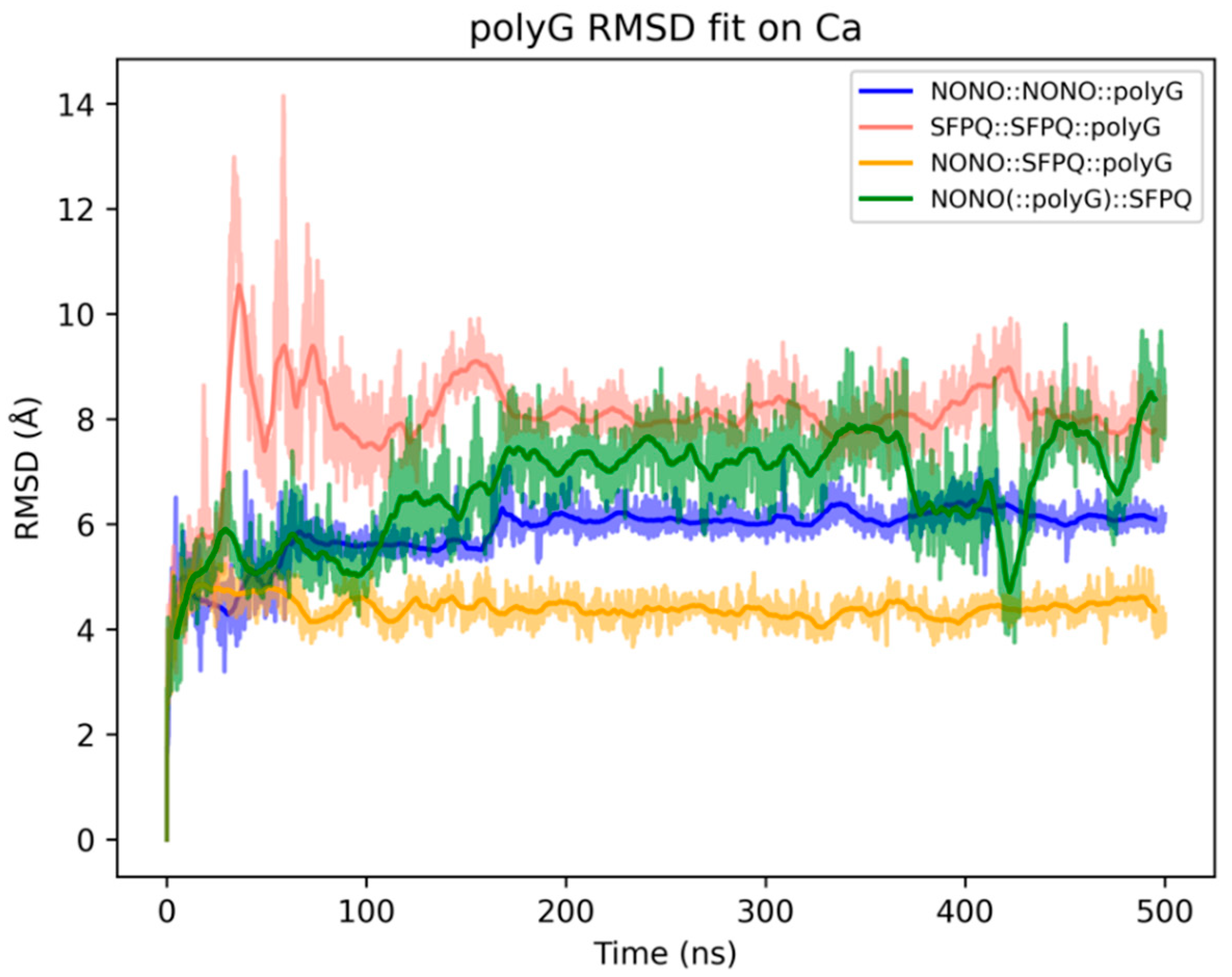
2.3. Analysis of NONO and SFPQ Dimer Interactions with RNA
3. Materials and Methods
3.1. Structure Preparation
3.2. Molecular Dynamics (MD)
3.3. Interactions Analysis
3.4. Dimer Formation Energy Calculation
3.5. In Silico Alanine Scan
3.6. DBS::PolyG Model Construction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.T.F.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENε/β Noncoding RNAs Are Essential for Structural Integrity of Nuclear Paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An Architectural Role for a Nuclear Noncoding RNA: NEAT1 RNA Is Essential for the Structure of Paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN ε/β Nuclear-Retained Non-Coding RNAs Are up-Regulated upon Muscle Differentiation and Are Essential Components of Paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.; Sadowska, A.; Bekere, I.; Ho, D.; Gully, B.S.; Lu, Y.; Iyer, K.S.; Trewhella, J.; Fox, A.H.; Bond, C.S. The Structure of Human SFPQ Reveals a Coiled-Coil Mediated Polymer Essential for Functional Aggregation in Gene Regulation. Nucleic Acids Res. 2015, 43, 3826–3840. [Google Scholar] [CrossRef]
- Kameoka, S.; Duque, P.; Konarska, M.M. P54nrb Associates with the 5′ Splice Site within Large Transcription/Splicing Complexes. EMBO J. 2004, 23, 1782–1791. [Google Scholar] [CrossRef] [Green Version]
- Emili, A.; Shales, M.; McCracken, S.; Xie, W.; Tucker, P.W.; Kobayashi, R.; Blencowe, B.J.; Ingles, C.J. Splicing and Transcription-Associated Proteins PSF and P54nrb/NonO Bind to the RNA Polymerase II CTD. RNA 2002, 8, 1102–1111. [Google Scholar] [CrossRef] [Green Version]
- Teigelkamp, S.; Mundt, C.; Achsel, T.; Will, C.L.; Lührmann, R. The Human U5 SnRNP-Specific 100-KD Protein Is an RS Domain-Containing, Putative RNA Helicase with Significant Homology to the Yeast Splicing Factor Prp28p. RNA 1997, 3, 1313–1326. [Google Scholar]
- Gozani, O.; Patton, J.G.; Reed, R. A Novel Set of Spliceosome-Associated Proteins and the Essential Splicing Factor PSF Bind Stably to Pre-MRNA Prior to Catalytic Step II of the Splicing Reaction. EMBO J. 1994, 13, 3356–3367. [Google Scholar] [CrossRef]
- Peng, R.; Dye, B.T.; Pérez, I.; Barnard, D.C.; Thompson, A.B.; Patton, J.G. PSF and P54nrb Bind a Conserved Stem in U5 SnRNA. RNA 2002, 8, 1334–1347. [Google Scholar] [CrossRef] [Green Version]
- Jaafar, L.; Li, Z.; Li, S.; Dynan, W.S. SFPQ•NONO and XLF Function Separately and Together to Promote DNA Double-Strand Break Repair via Canonical Nonhomologous End Joining. Nucleic Acids Res. 2017, 45, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-J.; Das, U.; Xie, W.; Ducasse, M.; Tucker, H.O. Altered Stoichiometry and Nuclear Delocalization of NonO and PSF Promote Cellular Senescence. Aging 2016, 8, 3356–3374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schell, B.; Legrand, P.; Ebastien Fribourg, S. Crystal Structure of SFPQ-NONO Heterodimer. Biochimie 2022, 198, 1–7. [Google Scholar] [CrossRef]
- Taiana, E.; Favasuli, V.; Ronchetti, D.; Todoerti, K.; Pelizzoni, F.; Manzoni, M.; Barbieri, M.; Fabris, S.; Silvestris, I.; Gallo Cantafio, M.E.; et al. Long Non-Coding RNA NEAT1 Targeting Impairs the DNA Repair Machinery and Triggers Anti-Tumor Activity in Multiple Myeloma. Leukemia 2020, 34, 234–244. [Google Scholar] [CrossRef]
- Major, A.T.; Hogarth, C.A.; Young, J.C.; Kurihara, Y.; Jans, D.A.; Loveland, K.L. Dynamic Paraspeckle Component Localisation during Spermatogenesis. Reproduction 2019, 158, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Bottini, S.; Hamouda-Tekaya, N.; Mategot, R.; Zaragosi, L.-E.; Audebert, S.; Pisano, S.; Grandjean, V.; Mauduit, C.; Benahmed, M.; Barbry, P.; et al. Post-Transcriptional Gene Silencing Mediated by MicroRNAs Is Controlled by Nucleoplasmic Sfpq. Nat. Commun. 2017, 8, 1189. [Google Scholar] [CrossRef] [PubMed]
- Nussbacher, J.K.; Tabet, R.; Yeo, G.W.; Lagier-Tourenne, C. Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 2019, 102, 294–320. [Google Scholar] [CrossRef]
- Knott, G.J.; Lee, M.; Passon, D.M.; Fox, A.H.; Bond, C.S. Caenorhabditis Elegans NONO-1: Insights into DBHS Protein Structure, Architecture, and Function. Protein Sci. 2015, 24, 2033–2043. [Google Scholar] [CrossRef] [Green Version]
- Passon, D.M.; Lee, M.; Rackham, O.; Stanley, W.A.; Sadowska, A.; Filipovska, A.; Fox, A.H.; Bond, C.S. Structure of the Heterodimer of Human NONO and Paraspeckle Protein Component 1 and Analysis of Its Role in Subnuclear Body Formation. Proc. Natl. Acad. Sci. USA 2012, 109, 4846–4850. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Casas Garcia, G.P.; Perugini, M.A.; Fox, A.H.; Bond, C.S.; Lee, M. Crystal Structure of a SFPQ/PSPC1 Heterodimer Provides Insights into Preferential Heterodimerization of Human DBHS Family Proteins. J. Biol. Chem. 2018, 293, 6593–6602. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.W.; Zhang, L.X.; Busch, R.K.; Farres, J.; Busch, H. Purification and Characterization of a DNA-Binding Heterodimer of 52 and 100 KDa from HeLa Cells. Biochem. J. 1993, 290, 267–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.; Lutz, C.S. P54nrb Is a Component of the SnRNP-Free U1A (SF-A) Complex That Promotes Pre-MRNA Cleavage during Polyadenylation. RNA 2006, 12, 111–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knott, G.J.; Chong, Y.S.; Passon, D.M.; Liang, X.; Deplazes, E.; Conte, M.R.; Marshall, A.C.; Lee, M.; Fox, A.H.; Bond, C.S. Structural Basis of Dimerization and Nucleic Acid Binding of Human DBHS Proteins NONO and PSPC1. Nucleic Acids Res. 2022, 50, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasa Murthy, U.M.; Rangarajan, P.N. Identification of Protein Interaction Regions of VINC/NEAT1/Men Epsilon RNA. FEBS Lett. 2010, 584, 1531–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teplova, M.; Song, J.; Gaw, H.Y.; Teplov, A.; Patel, D.J. Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1. Structure 2010, 18, 1364–1377. [Google Scholar] [CrossRef] [Green Version]
- Maris, C.; Dominguez, C.; Allain, F.H.T. The RNA Recognition Motif, a Plastic RNA-Binding Platform to Regulate Post-Transcriptional Gene Expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural LncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef] [Green Version]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of p K a Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, SC’06, New York, NY, USA, 11–17 November 2006. [Google Scholar]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Tina, K.G.; Bhadra, R.; Srinivasan, N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007, 35, W473–W476. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laurenzi, T.; Palazzolo, L.; Taiana, E.; Saporiti, S.; Ben Mariem, O.; Guerrini, U.; Neri, A.; Eberini, I. Molecular Modelling of NONO and SFPQ Dimerization Process and RNA Recognition Mechanism. Int. J. Mol. Sci. 2022, 23, 7626. https://doi.org/10.3390/ijms23147626
Laurenzi T, Palazzolo L, Taiana E, Saporiti S, Ben Mariem O, Guerrini U, Neri A, Eberini I. Molecular Modelling of NONO and SFPQ Dimerization Process and RNA Recognition Mechanism. International Journal of Molecular Sciences. 2022; 23(14):7626. https://doi.org/10.3390/ijms23147626
Chicago/Turabian StyleLaurenzi, Tommaso, Luca Palazzolo, Elisa Taiana, Simona Saporiti, Omar Ben Mariem, Uliano Guerrini, Antonino Neri, and Ivano Eberini. 2022. "Molecular Modelling of NONO and SFPQ Dimerization Process and RNA Recognition Mechanism" International Journal of Molecular Sciences 23, no. 14: 7626. https://doi.org/10.3390/ijms23147626





