The Roots of Rye (Secale cereale L.) Are Capable of Synthesizing Benzoxazinoids
Abstract
:1. Introduction
2. Results
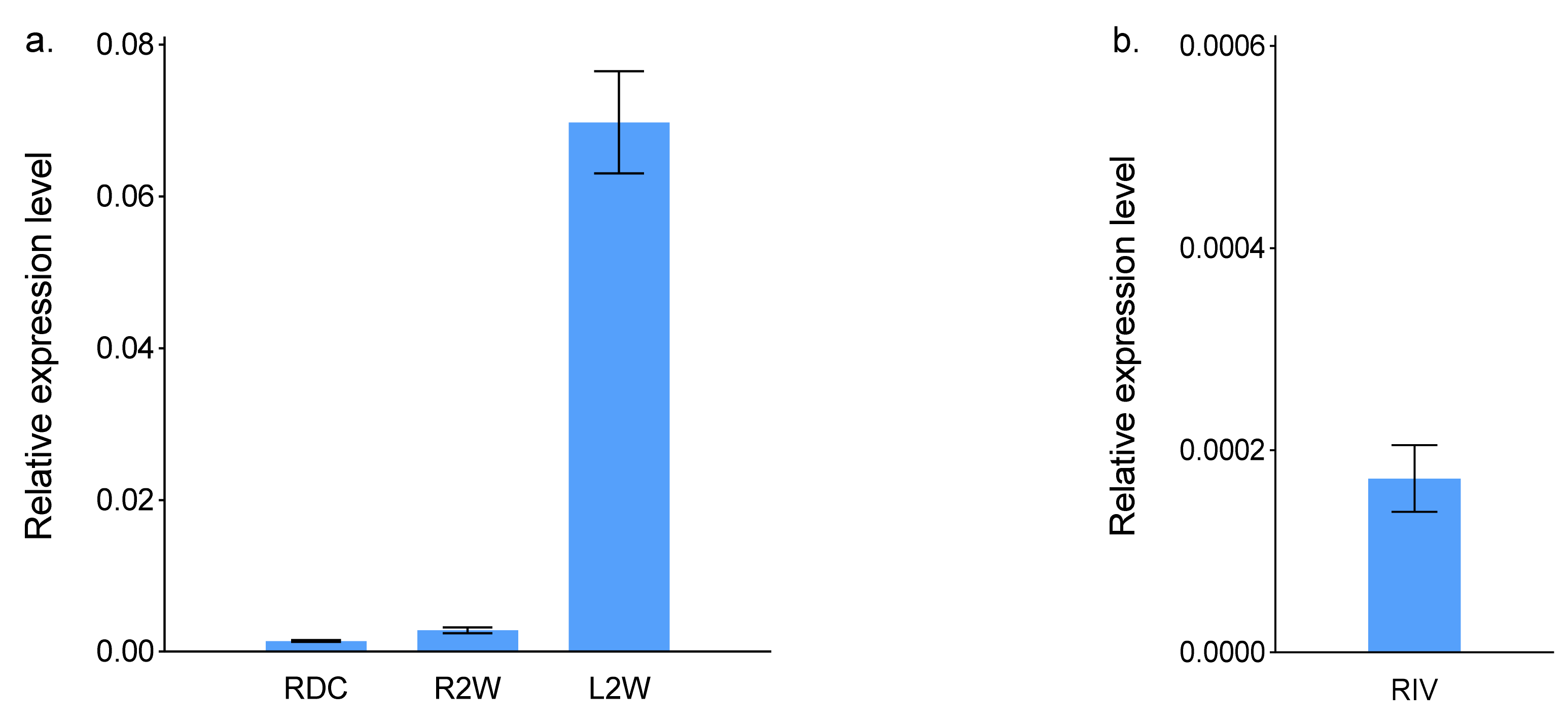
2.1. ScBx1 Gene Expression
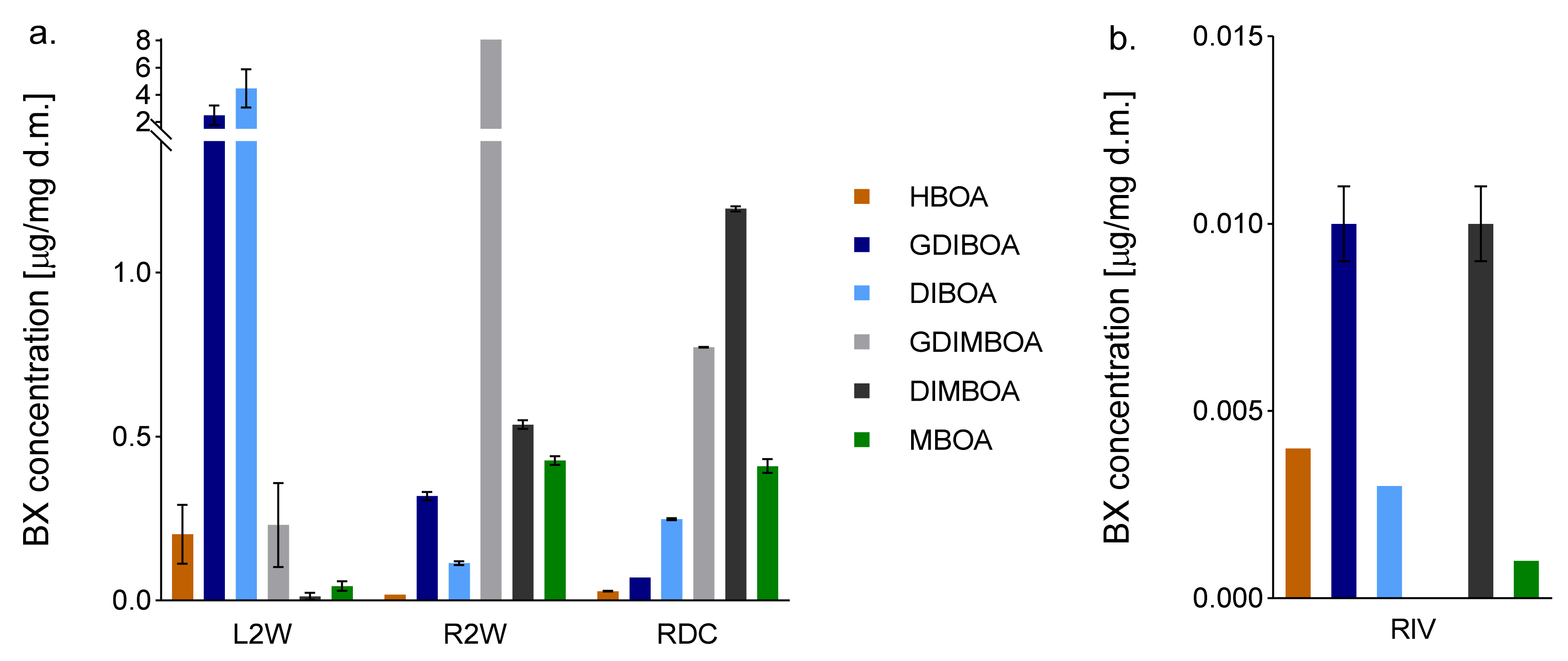
2.2. BX Content
2.3. Bioinformatic Characterisation of BX1 Protein
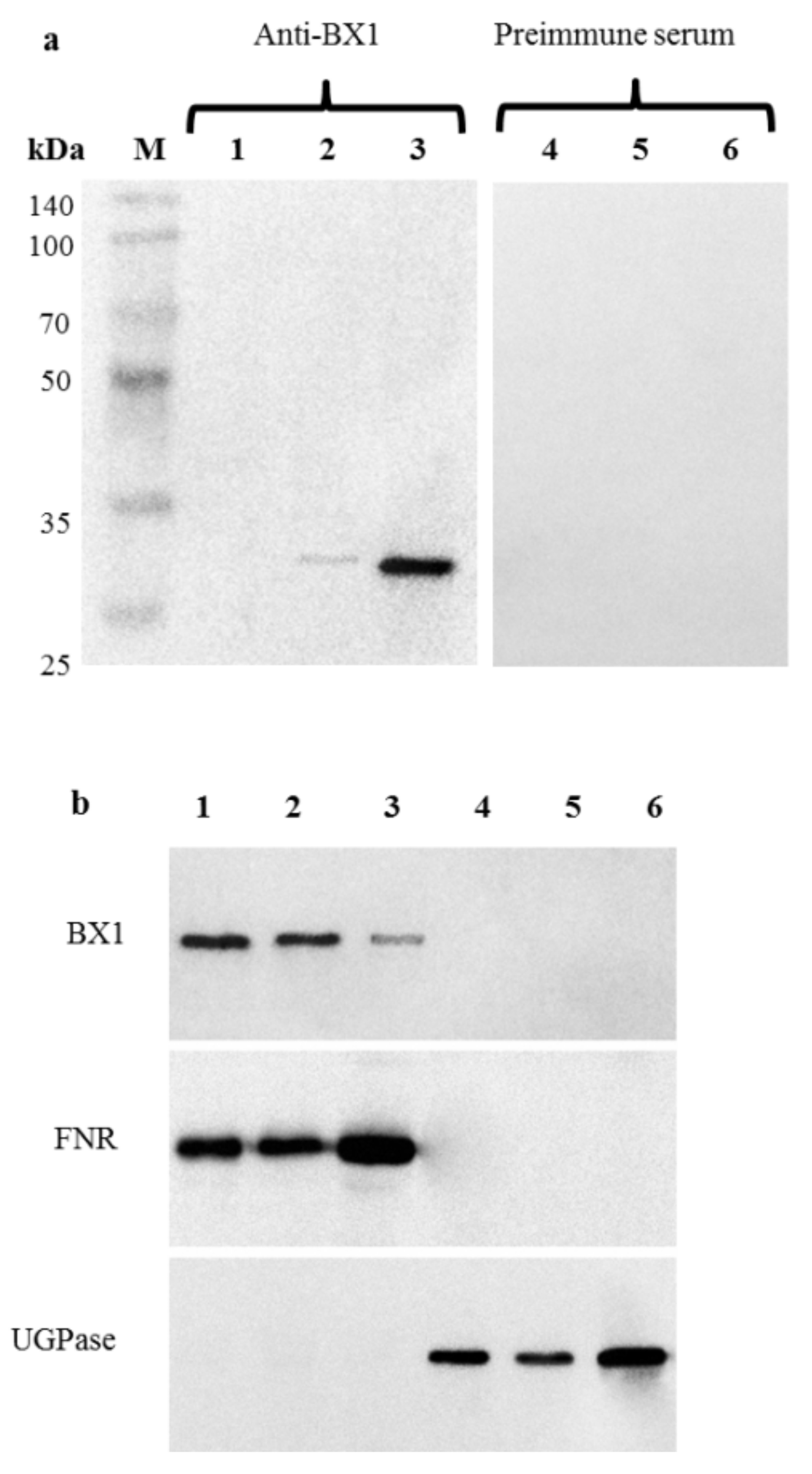
2.4. Immunodetection of BX1 in the Plastid Fraction of Roots
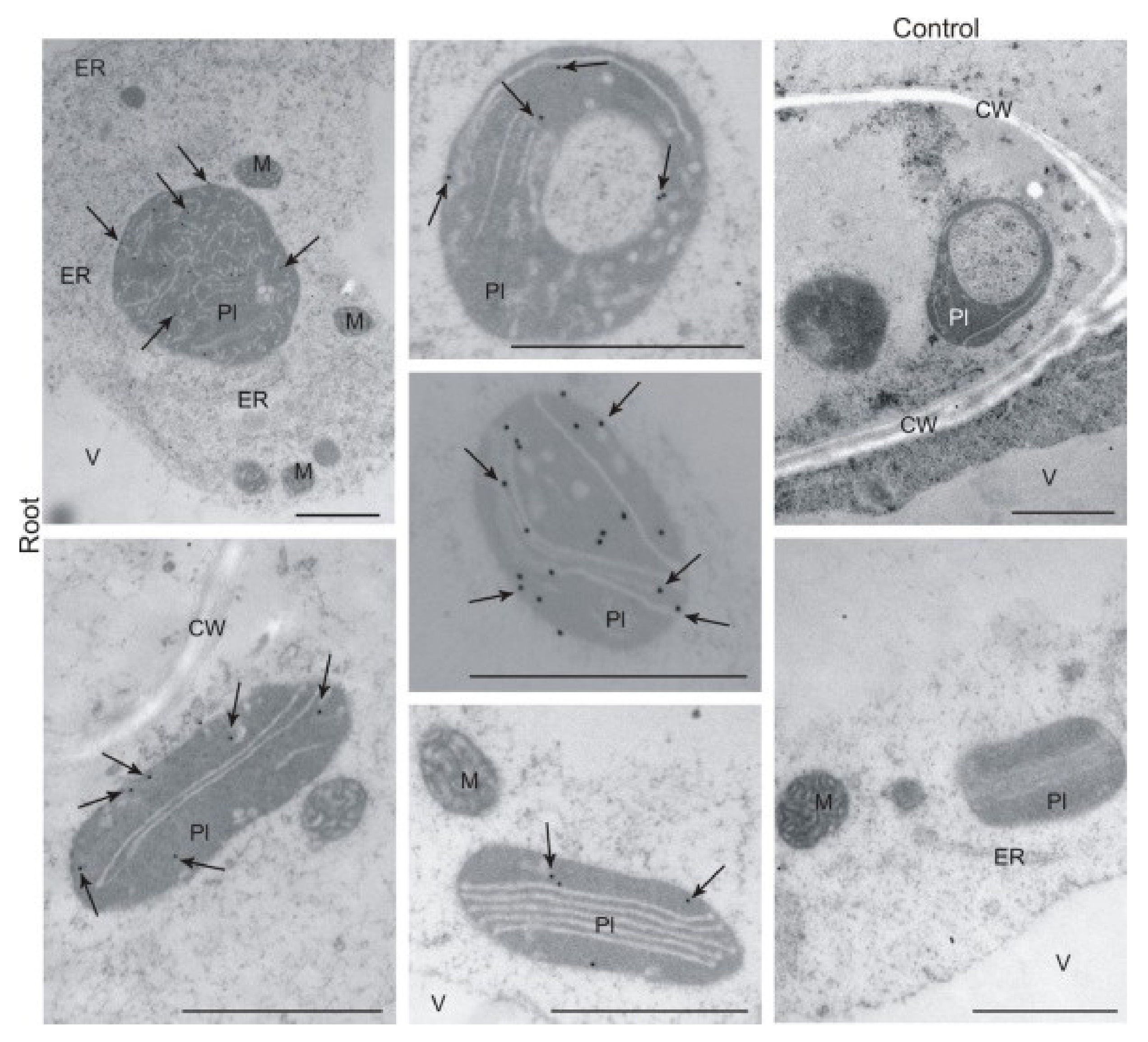
2.5. Immunolocalisation of BX1 Protein in Plastids of Roots and Leaves
3. Discussion
4. Materials and Methods
4.1. Plant Material
- -
- roots developed from seeds deprived of the coleoptile at 2 days after sowing (RDC),
- -
- roots developed in vitro (RIV),
- -
- roots (R2W) and leaves (L2W) from 2-week-old seedlings.
4.2. Bioinformatics Analysis of BX1
4.3. RNA Isolation, cDNA Synthesis, and Quantitative real Time-PCR Analysis
4.4. Biochemical Analysis
4.5. Immunodetection of BX1 Protein in Root and Leaf Plastids
4.5.1. Preparation of Antiserum
4.5.2. Plastid Isolation
4.5.3. Cytosol Isolation
4.5.4. Preparation of Protein Extracts
4.5.5. Western Blot Analysis
4.6. Detection of BX1 Protein by Transmission Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rice, C.P.; Park, Y.B.; Adam, F.; Abdul-Baki, A.A.; Teasdale, J.R. Hydroxamic acid content and toxicity of rye at selected growth stages. J. Chem. Ecol. 2005, 31, 1887–1905. [Google Scholar] [CrossRef]
- Schulz, M.; Marocco, A.; Tabaglio, V.; Macías, F.; Molinillo, J. Benzoxazinoids in rye allelopathy—from discovery to application in sustainable weed control and organic farming. J. Chem. Ecol. 2013, 39, 154–174. [Google Scholar] [CrossRef]
- Makowska, B.; Bakera, B.; Rakoczy-Trojanowska, M. The genetic background of benzoxazinoid biosynthesis in cereals. Acta Physiol. Plant. 2015, 37, 176. [Google Scholar] [CrossRef] [Green Version]
- Frey, M.; Schullehner, K.; Dick, R.; Fiesselmann, A.; Gierl, A. Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 2009, 70, 1645–1651. [Google Scholar] [CrossRef]
- Meihls, L.N.; Handrick, V.; Glauser, G.; Barbier, H.; Kaur, H.; Haribal, M.M.; Lipka, A.E.; Gershenzon, J.; Buckler, E.S.; Erb, M.; et al. Natural variation in maize aphid resistance is associated with 2, 4-dihydroxy-7-methoxy-1, 4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 2013, 25, 2341–2355. [Google Scholar] [CrossRef] [Green Version]
- Handrick, V.; Robert, C.A.M.; Ahern, K.R.; Zhou, S.; Machado, R.A.R.; Maag, D.; Glauser, G.; Fernandez-Penny, F.E.; Chandran, J.N.; Rodgers-Melnik, E.; et al. Biosynthesis of 8-O-methylated benzoxazinoid defence compounds in maize. Plant Cell 2016, 28, 1682–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wouters, F.C.; Blanchett, B.; Gershenzo, J.; Vassão, D.G. Plant defense and herbivore counter-defense: Benzoxazinoids and insect herbivores. Phytochem. Rev. 2016, 15, 1127–1151. [Google Scholar] [CrossRef] [Green Version]
- La Hovary, C. Allelochemicals in Secale cereale: Biosynthesis and Molecular Biology of Benzoxazinones. 2011. Available online: https://repository.lib.ncsu.edu/bitstream/handle/1840.16/6844/etd.pdf?sequence=2 (accessed on 20 January 2021).
- Bakera, B.; Makowska, B.; Groszyk, J.; Niziołek, M.; Orczyk, W.; Bolibok-Brągoszewska, H.; Hromada-Judycka, A.; Rakoczy-Trojanowska, M. Structural characteristics of ScBx genes controlling the biosynthesis of hydroxamic acids in rye (Secale cereale L.). J. Appl. Genet. 2016, 57, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanwir, F.; Dionisio, G.; Adhikari, K.B.; Fomsgaard, I.S.; Gregersen, P.L. Biosynthesis and chemical transformation of benzoxazinoids in rye during seed germination and the identification of a rye Bx6-like gene. Phytochemistry 2017, 140, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Wlazło, A.; Święcicka, M.; Koter, M.D.; Krępski, T.; Bolibok, L.; Stochmal, A.; Kowalczyk, M.; Rakoczy-Trojanowska, M. Genes ScBx1 and ScIgl–Competitors or Cooperators? Genes 2020, 11, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frey, M.; Chomet, P.; Glawischnig, E.; Stettner, C.; Grun, S.; Winklmair, A.; Eisenreich, W.; Bacher, A.; Meeley, R.B.; Briggs, S.P.; et al. Analysis of a chemical plant defense mechanism in grasses. Science 1997, 277, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Frey, M.; Stettner, C.; Pare, P.W.; Schmelz, E.A.; Tumlinson, J.H.; Gierl, A. An herbivore elicitor activates the gene for indole emission in maize. PNAS 2000, 97, 14801–14806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakoczy-Trojanowska, M.; Święcicka, M.; Bakera, B.; Kowalczyk, M.; Stochmal, A.; Bolibok, L. Co-cultivating rye with berseem clover affects benzoxazinoid production and expression of related genes. Crop Sci. 2020, 60, 3228–3246. [Google Scholar] [CrossRef]
- Rakoczy-Trojanowska, M.; Święcicka, M.; Rymuszka, J.; Stochmal, A.; Kowalczyk, M. New aspects of the genetic background of benzoxazinoid biosynthesis in rye Secale cereale L. In Proceedings of the EUCARPIA Cereal Section/IWIW2 Meetings, Polydôme, Clermont-Ferrand, France, 19–22 March 2018. [Google Scholar]
- Rakoczy-Trojanowska, M.; Święcicka, M.; Bakera, B.; Wlazło, A. Genetic and environmental determinants regulating benzoxazinoid biosynthesis in rye (Secale cereale L)—facts and myths. In Proceedings of the Integrative Plant Biology Conference, IPG PAS, Poznan, Poland, 7–9 November 2018. [Google Scholar]
- Nomura, T.; Ishihara, A.; Yanagita, R.C.; Endo, T.R.; Iwamura, H. Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. PNAS 2005, 102, 16490–16495. [Google Scholar] [CrossRef] [Green Version]
- Von Rad, U.; Huttl, R.; Lottspeich, F.; Gierl, A.; Frey, M. Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 2001, 28, 633–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Doan, C.; Züst, T.; Maurer, C.; Zhang, X.; Machado, R.A.R.; Mateo, P.; Ye, M.; Schimmel, B.C.J.; Glauser, G.; Robert, C.A.M. Tissue-specific volatile-mediated defense regulation in maize leaves and roots. bioRxiv 2020. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef] [Green Version]
- Neal, A.L.; Ton, J. Systemic defense priming by Pseudomonas putida KT2440 in maize depends on benzoxazinoid exudation from the roots. Plant Signal. Behav. 2013, 8, e22655. [Google Scholar] [CrossRef] [Green Version]
- Cotton, T.E.A.; Pétriacq, P.; Cameron, D.D.; Meselmani, M.A.; Schwarzenbacher, R.; Rolfe, S.A.; Tin, J. Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 2019, 13, 1647–1658. [Google Scholar] [CrossRef] [Green Version]
- Pérez, F.J.; Ormenoñuñez, J. Difference in hydroxamic acid content in roots and root exudates of wheat (Triticum aestivum L.) and rye (Secale cereale L.): Possible role in allelopathy. J. Chem. Ecol. 1991, 17, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Belz, R.G.; Hurle, K. Differential exudation of two benzoxazinoids—One of the determining factors for seedling allelopathy of Triticeae species. J. Agric. Food Chem. 2005, 53, 250–261. [Google Scholar] [CrossRef]
- Stochmal, A.; Kus, J.; Martyniuk, S.; Oleszek, W. Concentration of benzoxazinoids in roots of field-grown wheat (Triticum aestivum L.) varieties. J. Agric. Food Chem. 2006, 54, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Villagrasa, M.; Guillamón, M.; Labandeira, A.; Taberner, A.; Eljarrat, E.; Barceló, D. Benzoxazinoid allelochemicals in wheat: distribution among foliage, roots, and seeds. J. Agric. Food Chem. 2006, 54, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Ben-Abu, Y.; Beiles, A.; Flom, D.; Nevo, E. Adaptive evolution of benzoxazinoids in wild emmer wheat, Triticum dicoccoides, at “EvolutionCanyon”, MountCarmel, Israel. PLoS ONE 2018, 13, e0190424. [Google Scholar] [CrossRef] [Green Version]
- Zasada, I.A.; Rice, C.P.; Meyer, S.L.F. Improving the use of rye (Secale cereale) for nematode management: Potential to select cultivars based on Meloidogyne incognita host status and benzoxazinoid content. Nematology 2007, 9, 53–60. [Google Scholar] [CrossRef]
- Meyer, S.L.F.; Rice, C.P.; Zasada, I.A. DIBOA: Fate in soil and effects on root—Knot nematode egg numbers. Soil Biol. Biochem. 2009, 41, 1555–1560. [Google Scholar] [CrossRef]
- Rice, C.P.; Cai, G.; Teasdale, J.R. Concentrations and allelopathic effects of benzoxazinoid compounds in soil treated with rye (Secale cereale) cover crop. J. Agric. Food Chem. 2012, 60, 4471–4479. [Google Scholar] [CrossRef]
- Tabaglio, V.; Gavazzi, C.; Schulz, M.; Marocco, A. Alternative weed control using the allelopathic effect of natural benzoxazinoids from rye mulch. Agron. Sustain. Dev. 2008, 28, 397–401, hal-00886420. [Google Scholar] [CrossRef]
- Gavazzi, C.; Schulz, M.; Marocco, A.; Tabaglio, V. Sustainable weed control by allelochemicals from rye cover crops: From the greenhouse to field evidence. Allelopathy J. 2010, 25, 259–273. [Google Scholar]
- Givovich, A.; Sandström, J.; Niemeyer, H.M.; Pettersson, J. Presence of a hydroxamic acid glucoside in wheat phloem sap, and its consequences for performance of Rhopalosiphum padi (L.) (Homoptera: Aphididae). J. Chem. Ecol. 1994, 20, 1923–1930. [Google Scholar] [CrossRef]
- Kataoka, T.; Hayashi, N.; Yamaya, T.; Takahashi, H. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004, 136, 4198–4204. [Google Scholar] [CrossRef] [Green Version]
- Shimpei, U.; Shinsuke, M.; Masato, K.; Akira, K.; Tomohito, A.; Satoru, I. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J. Exp. Bot. 2009, 60, 2677–2688. [Google Scholar] [CrossRef] [Green Version]
- Ko, D.; Kang, J.; Kiba, T.; Park, J.; Kojima, M.; Do, J.; Kim, K.Y.; Kwon, M.; Endler, A.; Song, W.Y.; et al. Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. PNAS 2014, 111, 7150–7155. [Google Scholar] [CrossRef] [Green Version]
- Hazrati, H.; Fomsgaard, I.S.; Kudsk, P. Root-exuded benzoxazinoids: Uptake and translocation in neighboring plants. J. Agric. Food Chem. 2020, 68, 10609–10617. [Google Scholar] [CrossRef]
- Espinosa-Leal, C.A.; Puente-Garza, C.A.; García-Lara, S. In vitro plant tissue culture: Means for production of biological active compounds. Planta 2018, 248, 1–18. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.; Jang, M.O.; Jin, Y.W.; Lee, E.K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Gierl, A.; Frey, M. Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 2001, 213, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Fiesselmannb, A.; Zhaoa, N.; Chena, H.; Frey, M.; Chen, F. Biosynthesis and emission of insect herbivory-induced volatile indole in rice. Phytochemistry 2011, 73, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Bermúdez, I.C.; Schmidt, W. The conundrum of discordant protein and mRNA expression. Are plants special? Front. Plant Sci. 2014, 5, 616. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sablok, G.; Powell, J.J.; Kazan, K. Emerging Roles and Landscape of Translating mRNAs in Plants. Front. Plant Sci. 2017, 8, 1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echevarría-Zomeño, S.; Yángüez, E.; Fernández-Bautista, N.; Castro-Sanz, A.B.; Ferrando, A.; Castellano, M.M. Regulation of translation initiation under biotic and abiotic stresses. Int. J. Mol. Sci. 2013, 14, 4670–4683. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Lin, W.D.; Ray, P.; Lan, P.; Schmidt, W. Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiol. 2013, 162, 1750–1763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandran, H.; Meena, M.; Barupal, T.; Sharma, K. Plant tissue culture as a perpetual source for production of industrially important bioactive compounds. Biotechnol. Rep. 2020, 26, e00450. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.S.; Jiang, Q.T.; Ma, J.; Wang, X.Y.; Wang, J.R.; Chen, G.Y.; Qi, P.F.; Pen, Y.Y.; Lan, X.J.; Zheng, Y.L.; et al. Characterization and expression analyses of the H+-pyrophosphatase gene in rye. J. Genet. 2016, 95, 565–572. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Święcicka, M.; Dmochowska-Boguta, M.; Orczyk, W.; Grądzielewska, A.; Stochmal, A.; Kowalczyk, M.; Bolibok, L.; Rakoczy-Trojanowska, M. Changes in benzoxazinoid contents and the expression of the associated genes in rye (Secale cereale L.) due to brown rust and the inoculation procedure. PLoS ONE 2020, 15, e0233807. [Google Scholar] [CrossRef] [PubMed]
- Brorson, S.H.; Hansen, A.R.; Nielsen, H.Z.; Woxen, I.K. A comparative study of the immunogold labelling on H2O2—treated and heated epoxy sections. Micron 1999, 32, 147–151. [Google Scholar] [CrossRef]

| Gene | Sequences (5′–3′) |
|---|---|
| ScBx1 | F: TCAAAACCTGAACACGTGAAGC |
| R: GCCTCTAGCCTTTTCAATCCTTC | |
| HvAct | F: CCCCTTTGAACCCAAAAGCC |
| R: GAAAGCACGGCCTGAATAGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rakoczy-Trojanowska, M.; Szabała, B.M.; Różańska, E.; Kowalczyk, M.; Burza, W.; Bakera, B.; Święcicka, M. The Roots of Rye (Secale cereale L.) Are Capable of Synthesizing Benzoxazinoids. Int. J. Mol. Sci. 2021, 22, 4656. https://doi.org/10.3390/ijms22094656
Rakoczy-Trojanowska M, Szabała BM, Różańska E, Kowalczyk M, Burza W, Bakera B, Święcicka M. The Roots of Rye (Secale cereale L.) Are Capable of Synthesizing Benzoxazinoids. International Journal of Molecular Sciences. 2021; 22(9):4656. https://doi.org/10.3390/ijms22094656
Chicago/Turabian StyleRakoczy-Trojanowska, Monika, Bartosz M. Szabała, Elżbieta Różańska, Mariusz Kowalczyk, Wojciech Burza, Beata Bakera, and Magdalena Święcicka. 2021. "The Roots of Rye (Secale cereale L.) Are Capable of Synthesizing Benzoxazinoids" International Journal of Molecular Sciences 22, no. 9: 4656. https://doi.org/10.3390/ijms22094656






