Enigmatic Pilus-Like Endospore Appendages of Bacillus cereus Group Species
Abstract
:1. Introduction
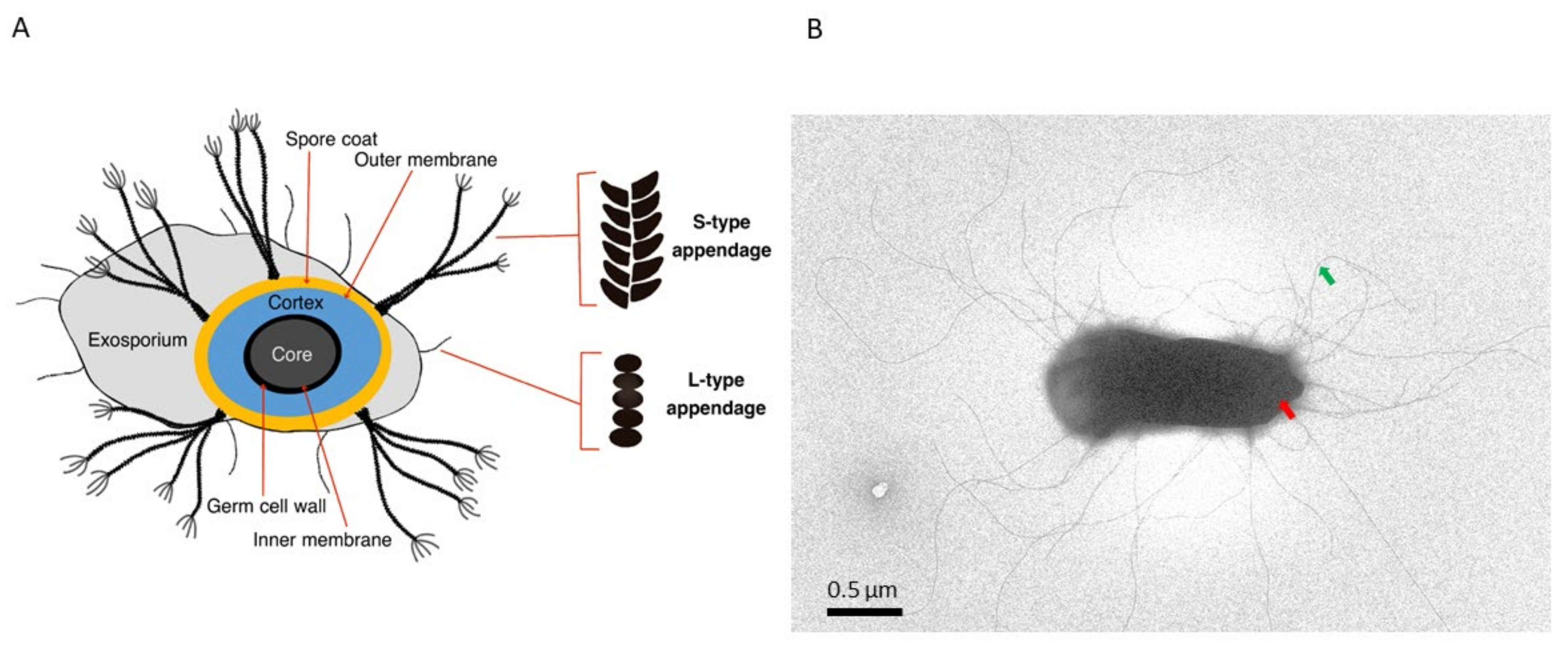
2. Bacillus Endospore Structure
3. Endospore Appendages
4. Architecture of Ena Fibers
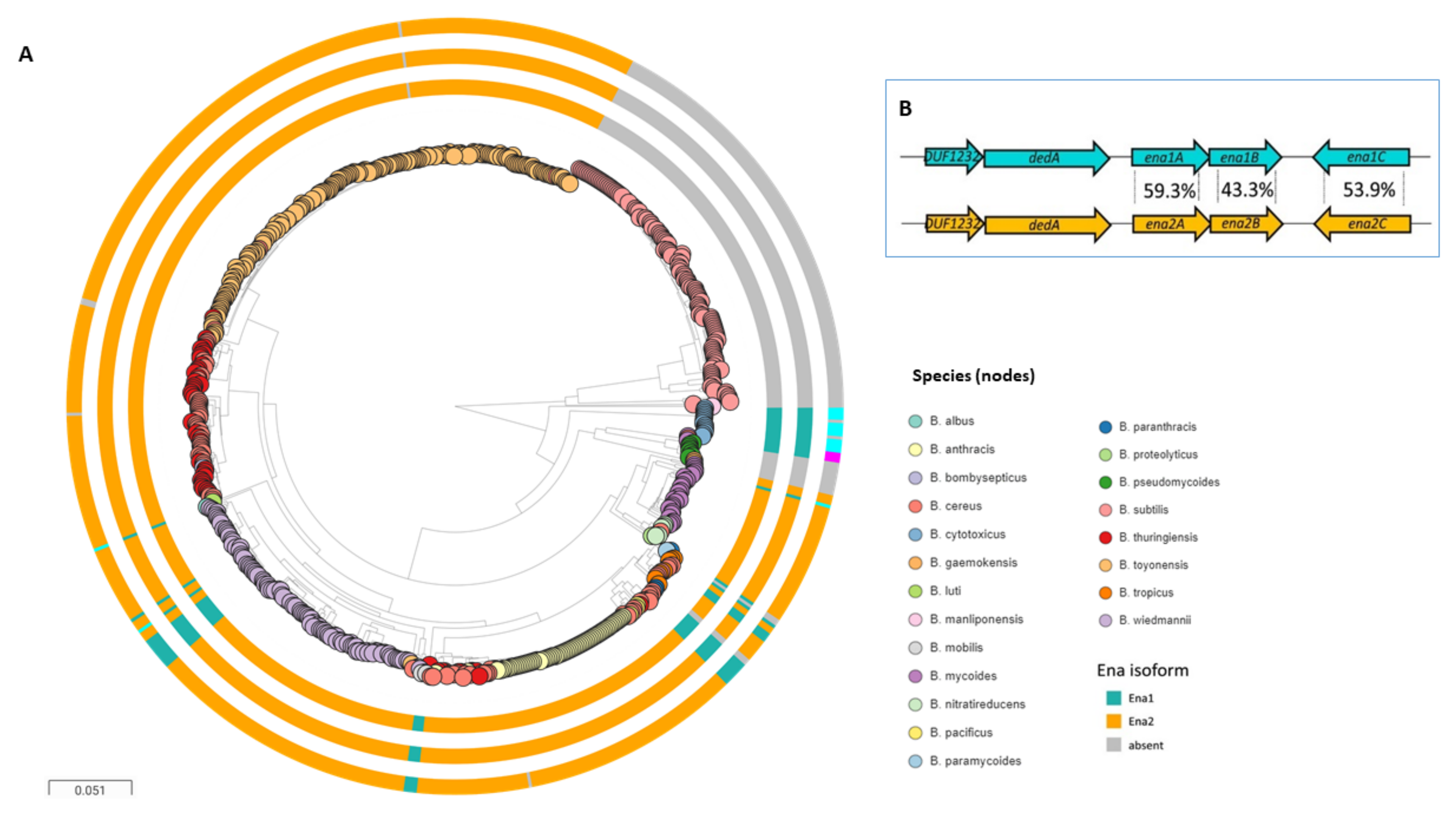
5. Ena-Encoding Genes
6. Distribution of Ena within the B. cereus s.l. Group
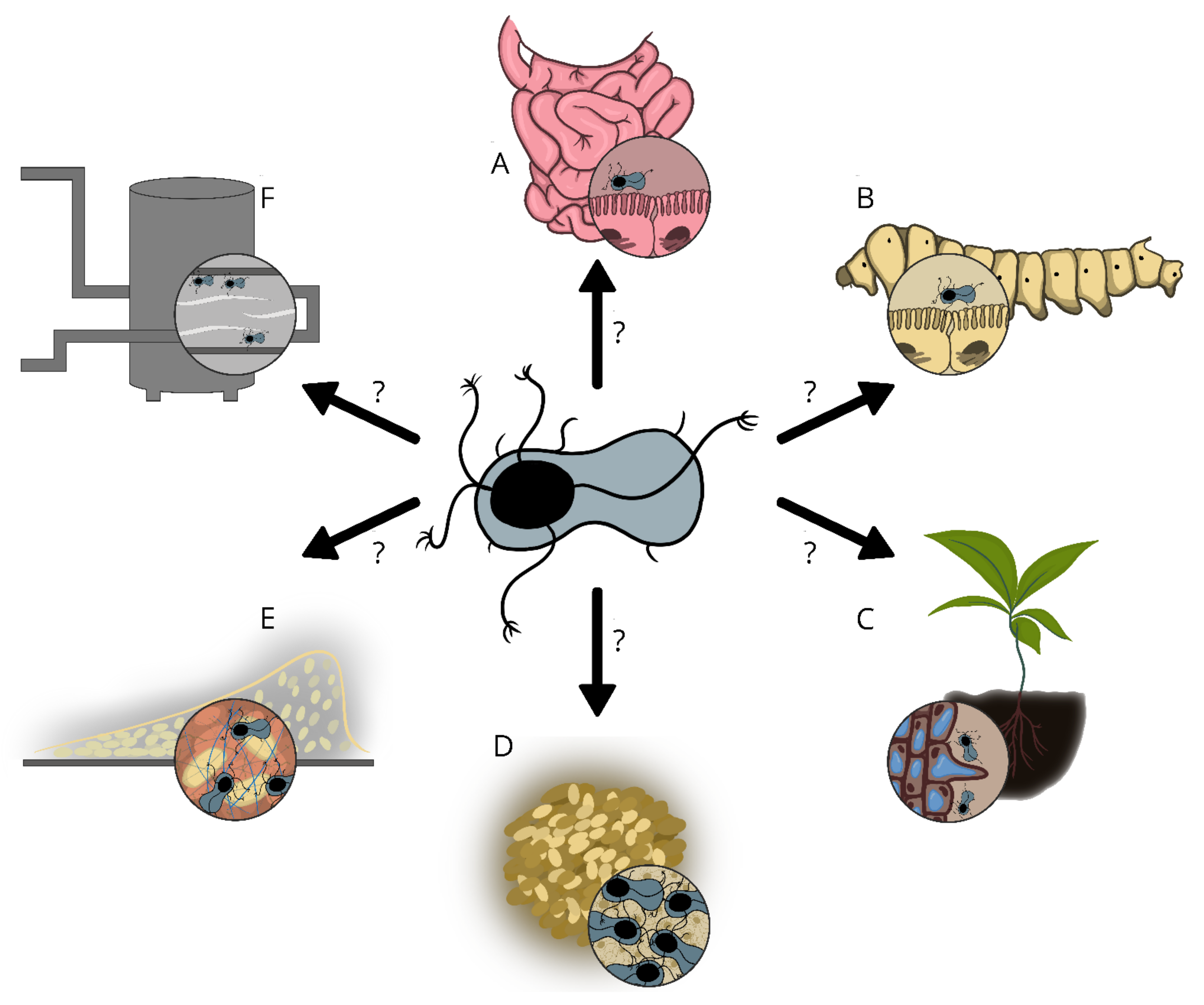
7. Potential Functions of Enas
8. Do Enas Function in Adhesion to Abiotic Surfaces?
9. Do Enas Promote the Adhesion of Endospores to Biotic Surfaces?
10. Enas in Autoaggregation and Biofilm Formation
11. Concluding Remarks
Funding
Acknowledgments
Conflicts of Interest
References
- Ramarao, N.; Lereclus, D. The InhA 1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell. Microbiol. 2005, 7, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Moir, A.; Cooper, G. Spore germination. Microbiol. Spectr. 2015, 3, 217–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klavenes, A.; Stalheim, T.; Sjøvold, O.; Josefsen, K.; Granum, P.E. Attachment of Bacillus cereus spores with and without appendages to stainless steel surfaces. Food Bioprod. Process. 2002, 80, 312–318. [Google Scholar] [CrossRef]
- Hayrapetyan, H.; Muller, L.; Tempelaars, M.; Abee, T.; Groot, M.N. Comparative analysis of biofilm formation by Bacillus cereus reference strains and undomesticated food isolates and the effect of free iron. Int. J. Food Microbiol. 2015, 200, 72–79. [Google Scholar] [CrossRef]
- Tauveron, G.; Slomianny, C.; Henry, C.; Faille, C. Variability among Bacillus cereus strains in spore surface properties and influence on their ability to contaminate food surface equipment. Int. J. Food Microbiol. 2006, 110, 254–262. [Google Scholar] [CrossRef]
- Faille, C.; Sylla, Y.; Le Gentil, C.; Bénézech, T.; Slomianny, C.; Lequette, Y. Viability and surface properties of spores subjected to a cleaning-in-place procedure: Consequences on their ability to contaminate surfaces of equipment. Food Microbiol. 2010, 27, 769–776. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Mossel, B.L.; Intaraphan, T.; Deeth, H.C. Heat resistance of Bacillus spores when adhered to stainless steel and its relationship to spore hydrophobicity. J. Food Prot. 2003, 66, 2070–2075. [Google Scholar] [CrossRef]
- Rubio, S.L.; Moldenhauer, J.E. Effect of rubber stopper composition, preservative pretreatment and rinse water temperature on the moist heat resistance of Bacillus stearothermophilus ATCC 12980. PDA J. Pharm. Sci. Technol. 1995, 49, 29–31. [Google Scholar] [PubMed]
- Majed, R.; Faille, C.; Kallassy, M.; Gohar, M. Bacillus cereus biofilms-same, only different. Front. Microbiol. 2016, 7, 1054. [Google Scholar] [CrossRef]
- Faille, C.; Bénézech, T.; Midelet-Bourdin, G.; Lequette, Y.; Clarisse, M.; Ronse, G.; Ronse, A.; Slomianny, C. Sporulation of Bacillus spp. within biofilms: A potential source of contamination in food processing environments. Food Microbiol. 2014, 40, 64–74. [Google Scholar] [CrossRef]
- Ryu, J.H.; Beuchat, L.R. Biofilm formation and sporulation by Bacillus cereus on a stainless steel surface and subsequent resistance of vegetative cells and spores to chlorine, chlorine dioxide, and a peroxyacetic acid-based sanitizer. J. Food Prot. 2005, 68, 2614–2622. [Google Scholar] [CrossRef] [PubMed]
- Pituch, H. Clostridium difficile is no longer just a nosocomial infection or an infection of adults. Int. J. Antimicrob. Agents 2009, 33, S42–S45. [Google Scholar] [CrossRef]
- Joshi, L.T.; Phillips, D.S.; Williams, C.F.; Alyousef, A.; Baillie, L. Contribution of spores to the ability of Clostridium difficile to adhere to surfaces. Appl. Environ. Microbiol. 2012, 78, 7671–7679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enoch, D.A.; Aliyu, S.H. Is Clostridium difficile infection still a problem for hospitals? CMAJ 2012, 184, 17–18. [Google Scholar] [CrossRef] [Green Version]
- Abee, T.; Kovács, Á.T.; Kuipers, O.P.; van der Veen, S. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 2011, 22, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Flint, S.H.; Palmer, J.S. Bacillus cereus spores and toxins–The potential role of biofilms. Food Microbiol. 2020, 90, 103493. [Google Scholar] [CrossRef] [PubMed]
- Deal, A.; Klein, D.; Lopolito, P.; Schwarz, J.S. Cleaning and disinfection of Bacillus cereus biofilm. PDA J. Pharm. Sci. Technol. 2016, 70, 208–217. [Google Scholar] [CrossRef]
- Driks, A.; Eichenberger, P. The spore coat. Microbiol. Spectr. 2016, 4, 179–200. [Google Scholar] [CrossRef]
- Mckenney, P.T.; Driks, A.; Eichenberger, P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013, 11, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Setlow, P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007, 15, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Moeller, R.; Setlow, P.; Horneck, G.; Berger, T.; Reitz, G.; Rettberg, P.; Doherty, A.J.; Okayasu, R.; Nicholson, W.L. Roles of the major, small, acid-soluble spore proteins and spore-specific and universal DNA repair mechanisms in resistance of Bacillus subtilis spores to ionizing radiation from X rays and high-energy charged-particle bombardment. J. Bacteriol. 2008, 190, 1134–1140. [Google Scholar] [CrossRef] [Green Version]
- Moeller, R.; Setlow, P.; Reitz, G.; Nicholson, W.L. Roles of small, acid-soluble spore proteins and core water content in survival of Bacillus subtilis spores exposed to environmental solar UV radiation. Appl. Environ. Microbiol. 2009, 75, 5202–5208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunde, E.P.; Setlow, P.; Hederstedt, L.; Halle, B. The physical state of water in bacterial spores. Proc. Natl. Acad. Sci. USA 2009, 106, 19334–19339. [Google Scholar] [CrossRef] [Green Version]
- McKenney, P.T.; Eichenberger, P. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol. Microbiol. 2012, 83, 245–260. [Google Scholar] [CrossRef] [Green Version]
- Ghebrehiwet, B.; Tantral, L.; Titmus, M.A.; Panessa-Warren, B.J.; Tortora, G.T.; Wong, S.S.; Warren, J.B. The exosporium of B. cereus contains a binding site for gC1qR/p33: Implication in spore attachment and/or entry. In Current Topics in Innate Immunity; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2007. [Google Scholar]
- Lequette, Y.; Garénaux, E.; Tauveron, G.; Dumez, S.; Perchat, S.; Slomianny, C.; Lereclus, D.; Guérardel, Y.; Faille, C. Role played by exosporium glycoproteins in the surface properties of Bacillus cereus spores and in their adhesion to stainless steel. Appl. Environ. Microbiol. 2011, 77, 4905–4911. [Google Scholar] [CrossRef] [Green Version]
- Stewart, G.C. The exosporium layer of bacterial spores: A connection to the environment and the infected host. Microbiol. Mol. Biol. Rev. 2015, 79, 437–457. [Google Scholar] [CrossRef] [Green Version]
- Plomp, M.; Leighton, T.J.; Wheeler, K.E.; Malkin, A.J. Architecture and high-resolution structure of Bacillus thuringiensis and Bacillus cereus spore coat surfaces. Langmuir 2005, 21, 7892–7898. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, B.; Liedtke, J.; Sleutel, M.; Lindbäck, T.; Zegeye, E.D.; O’Sullivan, K.; Llarena, A.; Brynildsrud, O.; Aspholm, M.; Remaut, H. Endospore Appendages: A novel pilus superfamily from the endospores of pathogenic Bacilli. EMBO J. 2021, 40, e106887. [Google Scholar] [CrossRef]
- Rode, L.J.; Crawford, M.A.; Williams, M.G. Clostridium spores with ribbon-like appendages. J. Bacteriol. 1967, 93, 1160–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hachisuka, Y.; Kuno, T. Filamentous appendages of Bacillus cereus spores. Jpn. J. Microbiol. 1976, 20, 555–558. [Google Scholar] [CrossRef]
- Ankolekar, C.; Labbé, R.G. Physical characteristics of spores of food-associated isolates of the Bacillus cereus group. Appl. Environ. Microbiol. 2010, 76, 982–984. [Google Scholar] [CrossRef] [Green Version]
- Hachisuka, Y.; Kozuka, S. A new test of differentiation of Bacillus cereus and Bacillus anthracis based on the existence of spore appendages. Microbiol. Immunol. 1981, 25, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Hachisuka, Y.; Kozuka, S.; Tsujikawa, M. Exosporia and appendages of spores of Bacillus species. Microbiol. Immunol. 1984, 28, 619–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budzik, J.M.; Poor, C.B.; Faull, K.F.; Whitelegge, J.P.; He, C.; Schneewind, O. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc. Natl. Acad. Sci. USA 2009, 106, 19992–19997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukaszczyk, M.; Pradhan, B.; Remaut, H. The biosynthesis and structures of bacterial pili. In Bacterial Cell Walls and Membranes; Subcellular Biochemistry; Springer: Cham, Switzerland, 2019; Volume 92. [Google Scholar]
- Walker, J.R.; Gnanam, A.J.; Blinkova, A.L.; Hermandson, M.J.; Karymov, M.A.; Lyubchenko, Y.L.; Graves, P.R.; Haystead, T.A.; Linse, K.D. Clostridium taeniosporum spore ribbon-like appendage structure, composition and genes. Mol. Microbiol. 2007, 63, 629–643. [Google Scholar] [CrossRef]
- Kozuka, S.; Tochikubo, K. Properties and origin of filamentous appendages on spores of Bacillus cereus. Microbiol. Immunol. 1985, 29, 21–37. [Google Scholar] [CrossRef]
- Stalheim, T.; Granum, P.E. Characterization of spore appendages from Bacillus cereus strains. J. Appl. Microbiol. 2001, 91, 839–845. [Google Scholar] [CrossRef] [Green Version]
- Kozuka, S. Fragmentation and solubilization of filamentous appendages of Bacillus cereus spores. Nippon. Saikingaku Zasshi. Jpn. J. Bacteriol. 1993, 8, 541–550. [Google Scholar] [CrossRef] [Green Version]
- Callaway, E. “It opens up a whole new universe”: Revolutionary microscopy technique sees individual atoms for first time. Nature 2020, 582, 156–157. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981, 34, 167–339. [Google Scholar] [CrossRef] [PubMed]
- Kailas, L.; Terry, C.; Abbott, N.; Taylor, R.; Mullin, N.; Tzokov, S.B.; Todd, S.J.; Wallace, B.A.; Hobbs, J.K.; Moir, A.; et al. Surface architecture of endospores of the Bacillus cereus/anthracis/ thuringiensis family at the subnanometer scale. Proc. Natl. Acad. Sci. USA 2011, 108, 16014–16019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hae, J.K.; Paterson, N.G.; Gaspar, A.H.; Hung, T.T.; Baker, E.N. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc. Natl. Acad. Sci. USA 2009, 106, 16967–16971. [Google Scholar] [CrossRef] [Green Version]
- Shaik, M.M.; Maccagni, A.; Tourcier, G.; Di Guilmi, A.M.; Dessen, A. Structural basis of pilus anchoring by the ancillary pilin RrgC of Streptococcus pneumoniae. J. Biol. Chem. 2014, 289, 16988–16997. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Devarajan, B.; Reardon, M.E.; Dwivedi, P.; Krishnan, V.; Cisar, J.O.; Das, A.; Narayana, S.V.L.; Ton-That, H. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol. Microbiol. 2011, 81, 1205–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persson, K.; Esberg, A.; Claesson, R.; Strömberg, N. The pilin protein FimP from Actinomyces oris: Crystal structure and sequence analyses. PLoS ONE 2012, 7, e48364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deivanayagam, C.C.S.; Rich, R.L.; Carson, M.; Owens, R.T.; Danthuluri, S.; Bice, T.; Höök, M.; Narayana, S.V.L. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 2000, 8, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Hartman, R.; Eilers, B.J.; Bollschweiler, D.; Munson-McGee, J.H.; Engelhardt, H.; Young, M.J.; Lawrence, C.M. The molecular mechanism of cellular attachment for an Archaeal Virus. Structure 2019, 27, 1634–1646. [Google Scholar] [CrossRef]
- Khare, B.; Narayana, S.V.L. Pilus biogenesis of Gram-positive bacteria: Roles of sortases and implications for assembly. Protein Sci. 2017, 26, 1458–1473. [Google Scholar] [CrossRef]
- Kang, H.J.; Baker, E.N. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J. Biol. Chem. 2009, 284, 20729–20737. [Google Scholar] [CrossRef] [Green Version]
- Katz, L.; Griswold, T.; Morrison, S.; Caravas, J.; Zhang, S.; Bakker, H.; Deng, X.; Carleton, H. Mashtree: A rapid comparison of whole genome sequence files. J. Open Source Softw. 2019, 4, 1762. [Google Scholar] [CrossRef] [Green Version]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swick, M.C.; Koehler, T.M.; Driks, A. Surviving between hosts: Sporulation and transmission. Microbiol. Spectr. 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, O.E.; Sauer, K. Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 2012, 194, 2413–2425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, A.; Granum, P.E.; Rönner, U. The adhesion of Bacillus cereus spores to epithelial cells might be an additional virulence mechanism. Int. J. Food Microbiol. 1998, 39, 93–99. [Google Scholar] [CrossRef]
- Ramarao, N.; Lereclus, D. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect. 2006, 8, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Fälker, S.; Dahlberg, S.; Normark, S.; Henriques-Normark, B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 2009, 5, 580–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ton-That, H.; Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003, 50, 1429–1438. [Google Scholar] [CrossRef]
- Izoré, T.; Contreras-Martel, C.; El Mortaji, L.; Manzano, C.; Terrasse, R.; Vernet, T.; Di Guilmi, A.M.; Dessen, A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 2010, 18, 106–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Mishra, A.; Yang, J.; Cisar, J.O.; Das, A.; Ton-That, H. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J. Bacteriol. 2011, 193, 3197–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faille, C.; Jullien, C.; Fontaine, F.; Bellon-Fontaine, M.N.; Slomianny, C.; Benezech, T. Adhesion of Bacillus spores and Escherichia coli cells to inert surfaces: Role of surface hydrophobicity. Can. J. Microbiol. 2002, 48, 728–738. [Google Scholar] [CrossRef]
- Rönner, U.; Husmark, U.; Henriksson, A. Adhesion of Bacillus spores in relation to hydrophobicity. J. Appl. Bacteriol. 1990, 69, 550–556. [Google Scholar] [CrossRef]
- Van Loosdrecht, M.C.M.; Lyklema, J.; Norde, W.; Zehnder, A.J.B. Bacterial adhesion: A physicochemical approach. Microb. Ecol. 1989, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Husmark, U.; Rönner, U. The influence of hydrophobic, electrostatic and morphologic properties on the adhesion of Bacillus spores. Biofouling 1992, 5, 335–344. [Google Scholar] [CrossRef]
- Faille, C.; Tauveron, G.; Le Gentil-Lelièvre, C.; Slomianny, C. Occurrence of Bacillus cereus spores with a damaged exosporium: Consequences on the spore adhesion on surfaces of food processing lines. J. Food Prot. 2007, 70, 2346–2353. [Google Scholar] [CrossRef] [PubMed]
- Mercier-Bonin, M.; Dehouche, A.; Morchain, J.; Schmitz, P. Orientation and detachment dynamics of Bacillus spores from stainless steel under controlled shear flow: Modelling of the adhesion force. Int. J. Food Microbiol. 2011, 146, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Koshikawa, T.; Yamazaki, M.; Yoshimi, M.; Ogawa, S.; Yamada, A.; Watabe, K.; Torii, M. Surface hydrophobicity of spores of Bacillus spp. J. Gen. Microbiol. 1989, 135, 2717–2722. [Google Scholar] [CrossRef] [Green Version]
- Ehling-Schulz, M.; Fricker, M.; Scherer, S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol. Nutr. Food Res. 2004, 48, 479–487. [Google Scholar] [CrossRef]
- Tewari, A.; Abdullah, S. Bacillus cereus food poisoning: International and Indian perspective. J. Food Sci. Technol. 2015, 52, 2500–2511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granum, P.E.; Lund, T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997, 157, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Ceuppens, S.; Uyttendaele, M.; Drieskens, K.; Heyndrickx, M.; Rajkovic, A.; Boon, N.; Van de Wiele, T. Survival and germination of Bacillus cereus spores without outgrowth or enterotoxin production during in vitro simulation of gastrointestinal transit. Appl. Environ. Microbiol. 2012, 78, 7698–7705. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; Arias, S.; Chaignepain, S.; Denayrolles, M.; Schmitter, J.M.; Bressollier, P.; Urdaci, M.C. Identification of surface proteins involved in the adhesion of a probiotic Bacillus cereus strain to mucin and fibronectin. Microbiology 2009, 155, 1708–1716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijnands, L.M.; Dufrenne, J.B.; Van Leusden, F.M.; Abee, T. Germination of Bacillus cereus spores is induced by germinants from differentiated Caco-2 cells, a human cell line mimicking the epithelial cells of the small intestine. Appl. Environ. Microbiol. 2007, 73, 5052–5054. [Google Scholar] [CrossRef] [Green Version]
- Jouzani, G.S.; Valijanian, E.; Sharafi, R. Bacillus thuringiensis: A successful insecticide with new environmental features and tidings. Appl. Microbiol. Biotechnol. 2017, 101, 2691–2711. [Google Scholar] [CrossRef] [PubMed]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Narisawa, N.; Watanabe, T.; Kawarai, T.; Myozen, K.; Okazaki, S.; Ogihara, H.; Yamasaki, M. Formation of the spore clumps during heat treatment increases the heat resistance of bacterial spores. Int. J. Food Microbiol. 2005, 102, 107–111. [Google Scholar] [CrossRef]
- Aiba, S.; Toda, K. Some analysis of thermal inactivation of bacterial-spore clump. Ferm. Technol. 1966, 44, 301–304. [Google Scholar]
- Mamane, H.; Linden, K.G. Impact of particle aggregated microbes on UV disinfection. I: Evaluation of spore–clay aggregates and suspended Spores. J. Environ. Eng. 2006, 132, 596–606. [Google Scholar] [CrossRef]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Cao, Y.; Yu, L.; Tao, Y.; Zhou, Y.; Zhi, Q.; Lin, H. Characteristics and influencing factors of amyloid fibers in S. mutans biofilm. AMB Express 2019, 9, 31. [Google Scholar] [CrossRef] [Green Version]
- Caro-Astorga, J.; Pérez-García, A.; de Vicente, A.; Romero, D. A genomic region involved in the formation of adhesin fibers in Bacillus cereus biofilms. Front. Microbiol. 2014, 5, 745. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zegeye, E.D.; Pradhan, B.; Llarena, A.-K.; Aspholm, M. Enigmatic Pilus-Like Endospore Appendages of Bacillus cereus Group Species. Int. J. Mol. Sci. 2021, 22, 12367. https://doi.org/10.3390/ijms222212367
Zegeye ED, Pradhan B, Llarena A-K, Aspholm M. Enigmatic Pilus-Like Endospore Appendages of Bacillus cereus Group Species. International Journal of Molecular Sciences. 2021; 22(22):12367. https://doi.org/10.3390/ijms222212367
Chicago/Turabian StyleZegeye, Ephrem Debebe, Brajabandhu Pradhan, Ann-Katrin Llarena, and Marina Aspholm. 2021. "Enigmatic Pilus-Like Endospore Appendages of Bacillus cereus Group Species" International Journal of Molecular Sciences 22, no. 22: 12367. https://doi.org/10.3390/ijms222212367





