2-(2-Methyl-2-nitrovinyl)furan but Not Furvina Interfere with Staphylococcus aureus Agr Quorum-Sensing System and Potentiate the Action of Fusidic Acid against Biofilms
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Synthesis of Furvina and 2-Nitrovinylfuran Derivatives
4.2. Preparation of 2-Nitrovinylfuran Derivatives Stock Solutions
4.3. Microorganisms and Culture Conditions
4.4. Determination of the Minimum Inhibitory Concentration (MIC)
4.5. Detection of Quorum Sensing Inhibition
4.6. Molecular Docking
4.7. Effect of Furvina and 2-Nitrovinylfuran Derivatives on the Prevention of Biofilm Formation
4.8. Effect of Furvina and the Best QSI Candidate on Biofilm Susceptibility to Fusidic Acid
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QS | Quorum sensing |
| QSI | Quorum sensing inhibitor |
| SSTI | Skin and soft tissue infections |
| Ai | Autoinducers |
| HSL | N-acyl-homoserine lactone |
| AIP | Autoinducer peptides |
| Agr | Accessory gene regulator |
| EPS | Extracellular polymeric substances |
| MIC | Minimum inhibitory concentration |
| HCT | Highest concentration tested |
| DMSO | Dimethyl sulfoxide |
| TSB | Tryptic soy broth |
| Gfp | Green fluorescence protein |
| OD | Optical density |
| PS | Polystyrene |
| CV | Crystal violet |
| PBS | Phosphate buffer saline |
| PI | Propidium iodide |
References
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williamson, D.A.; Carter, G.P.; Howde, B.P. Current and Emerging Topical Antibacterials and Antiseptics: Agents, Action, and Resistance Patterns. Clin. Microbiol. Rev. 2017, 30, 827–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, C.M.; Bassler, B.L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, K.Y.; Otto, M. Quorum-sensing regulation in staphylococci—An overview. Front. Microbiol. 2015, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Eickhoff, M.J.; Bassler, B.L. SnapShot: Bacterial Quorum Sensing. Cell 2018, 174, 1328.e1. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Geisinger, E. Quorum Sensing in Staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [Green Version]
- Paramanantham, P.; Pattnaik, S.; Busi, S. Quorum Sensing in Bacterial Pathogenesis and Virulence. In Implication of Quorum Sensing System in Biofilm Formation and Virulence; Springer: Berlin/Heidelberg, Germany, 2018; pp. 111–132. [Google Scholar]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [Green Version]
- Quave, C.L.; Horswill, A.R. Flipping the switch: Tools for detecting small molecule inhibitors of staphylococcal virulence. Front. Microbiol. 2014, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Otto, M. Staphylococcal Infections: Mechanisms of Biofilm Maturation and Detachment as Critical Determinants of Pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef]
- Simões, M.; Bennett, R.N.; Rosa, E.A.S. Understanding antimicrobial activities of phytochemicals against multidrug resistant bacteria and biofilms. Nat. Prod. Rep. 2009, 26, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Simões, M. Antimicrobial Strategies Effective Against Infectious Bacterial Biofilms. Curr. Med. Chem. 2011, 18, 2129–2145. [Google Scholar] [CrossRef] [PubMed]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Saenz, H.L.; Götz, F.; Otto, M. Impact of the agr Quorum-Sensing System on Adherence to Polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000, 182, 1688–1693. [Google Scholar] [CrossRef] [Green Version]
- Hentzer, M.; Givskov, M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1300–1307. [Google Scholar] [CrossRef]
- Balamurugan, P.; Praveen Krishna, V.; Bharath, D.; Lavanya, R.; Vairaprakash, P.; Adline Princy, S. Staphylococcus aureus quorum regulator SarA targeted compound, 2-[(Methylamino)methyl]phenol inhibits biofilm and down-regulates virulence genes. Front. Microbiol. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Brackman, G.; Coenye, T. Quorum Sensing Inhibitors as Anti-Biofilm Agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef]
- Singh, N.; Romero, M.; Travanut, A.; Monteiro, P.F.; Jordana-Lluch, E.; Hardie, K.R.; Williams, P.; Alexander, M.R.; Alexander, C. Dual bioresponsive antibiotic and quorum sensing inhibitor combination nanoparticles for treatment of Pseudomonas aeruginosa biofilms in vitro and ex vivo. Biomater. Sci. 2019, 7, 4099–4111. [Google Scholar] [CrossRef] [Green Version]
- Brackman, G.; Breyne, K.; De Rycke, R.; Vermote, A.; Van Nieuwerburgh, F.; Meyer, E.; Van Calenbergh, S.; Coenye, T. The Quorum Sensing Inhibitor Hamamelitannin Increases Antibiotic Susceptibility of Staphylococcus aureus Biofilms by Affecting Peptidoglycan Biosynthesis and eDNA Release. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Sekhon, B.S. Repositioning drugs and biologics: Retargeting old/ existing drugs for potential new therapeutic applications. J. Pharm. Educ. Res. 2013, 4, 1–15. [Google Scholar]
- Parvathaneni, V.; Kulkarni, N.S.; Muth, A.; Gupta, V. Drug repurposing: A promising tool to accelerate the drug discovery process. Drug Discov. Today 2019, 24, 2076–2085. [Google Scholar] [CrossRef] [PubMed]
- Fabbretti, A.; Brandi, L.; Petrelli, D.; Pon, C.L.; Castañedo, N.R.; Medina, R.; Gualerzi, C.O. The antibiotic Furvina® targets the P-site of 30S ribosomal subunits and inhibits translation initiation displaying start codon bias. Nucleic Acids Res. 2012, 40, 10366–10374. [Google Scholar] [CrossRef] [PubMed]
- Fabbretti, A.O.; Gualerzi, C.; Brandi, L. Comparative analysis of the mechanism of action of Furvina® and GE81112, two translation initiation inhibitors. In Worldwide Research Efforts in the Fighting Against Microbial Pathogens; Brown Walker Press: Irvine, CA, USA, 2013; pp. 114–117. ISBN 1612336361. [Google Scholar]
- Borges, A.; Sousa, P.; Gaspar, A.; Vilar, S.; Borges, F.; Simões, M. Furvina inhibits the 3-oxo-C12-HSL-based quorum sensing system of Pseudomonas aeruginosa and QS-dependent phenotypes. Biofouling 2017, 33, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective Chemical Inhibition of agr Quorum Sensing in Staphylococcus aureus Promotes Host Defense with Minimal Impact on Resistance. PLoS Pathog. 2014, 10, e1004174. [Google Scholar] [CrossRef] [PubMed]
- Leonard, P.G.; Bezar, I.F.; Sidote, D.J.; Stock, A.M. Identification of a hydrophobic cleft in the LytTR domain of AgrA as a locus for small molecule interactions that inhibit DNA binding. Biochemistry 2012, 51, 10035–10043. [Google Scholar] [CrossRef] [Green Version]
- Sidote, D.J.; Barbieri, C.M.; Wu, T.; Stock, A.M. Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with an Unusual Mode of Binding. Structure 2008, 16, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Rajasree, K.; Fasim, A.; Gopal, B. Conformational features of the Staphylococcus aureus AgrA-promoter interactions rationalize quorum-sensing triggered gene expression. Biochem. Biophys. Rep. 2016, 6, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Schillaci, D.; Spanò, V.; Parrino, B.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G.; Cascioferro, S. Pharmaceutical Approaches to Target Antibiotic Resistance Mechanisms. J. Med. Chem. 2017, 60, 8268–8297. [Google Scholar] [CrossRef]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrino, B.; Schillaci, D.; Carnevale, I.; Giovannetti, E.; Diana, P.; Cirrincione, G.; Cascioferro, S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019, 161, 154–178. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Swamy, S.S.; Kaur, G.; Princy, S.A.; Balamurugan, P. Synergism between quorum sensing inhibitors and antibiotics: Combating the antibiotic resistance crisis. In Biotechnological Applications of Quorum Sensing Inhibitors; Springer: Singapore, 2018; pp. 209–225. ISBN 9789811090264. [Google Scholar]
- Vicente-Soler, J.; Madrid, M.; Franco, A.; Soto, T.; Cansado, J.; Gacto, M. Quorum Sensing as Target for Antimicrobial Chemotherapy Jerónima. In New Weapons to Control Bacterial Growth; Springer: Singapore, 2016; pp. 161–184. ISBN 9783319283685. [Google Scholar]
- Scholz, T.; Heyl, C.L.; Bernardi, D.; Zimmermann, S.; Kattner, L.; Klein, C.D. Chemical, biochemical and microbiological properties of a brominated nitrovinylfuran with broad-spectrum antibacterial activity. Bioorg. Med. Chem. 2013, 21, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Milhazes, N.; Calheiros, R.; Marques, M.P.M.; Garrido, J.; Cordeiro, M.N.D.S.; Rodrigues, C.; Quinteira, S.; Novais, C.; Peixe, L.; Borges, F. β-Nitrostyrene derivatives as potential antibacterial agents: A structure-property-activity relationship study. Bioorg. Med. Chem. 2006, 14, 4078–4088. [Google Scholar] [CrossRef] [Green Version]
- Allas, Ü.; Toom, L.; Selyutina, A.; Mäeorg, U.; Medina, R.; Merits, A.; Rinken, A.; Hauryliuk, V.; Kaldalu, N.; Tenson, T. Antibacterial activity of the nitrovinylfuran G1 (Furvina) and its conversion products. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Borges, A.; Abreu, A.C.; Dias, C.; Saavedra, M.J.; Borges, F.; Simões, M. New perspectives on the use of phytochemicals as an emergent strategy to control bacterial infections including biofilms. Molecules 2016, 21, 877. [Google Scholar] [CrossRef]
- Chen, Y.; Koripella, R.K.; Sanyal, S.; Selmer, M. Staphylococcus aureus elongation factor G—Structure and analysis of a target for fusidic acid. FEBS J. 2010, 277, 3789–3803. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, P. Fusidic acid: A bacterial elongation factor inhibitor for the oral treatment of acute and chronic staphylococcal infections. Cold Spring Harb. Perspect. Med. 2016, 6, 1–18. [Google Scholar] [CrossRef]
- Shopsin, B.; Eaton, C.; Wasserman, G.A.; Mathema, B.; Adhikari, R.P.; Agolory, S.; Altman, D.R.; Holzman, R.S.; Kreiswirth, B.N.; Novick, R.P. Mutations in agr Do Not Persist in Natural Populations of Methicillin-Resistant Staphylococcus aureus. J. Infect. Dis. 2010, 202, 1593–1599. [Google Scholar] [CrossRef] [Green Version]
- Smyth, D.S.; Kafer, J.M.; Wasserman, G.A.; Velickovic, L.; Mathema, B.; Holzman, R.S.; Knipe, T.A.; Becker, K.; Von Eiff, C.; Peters, G.; et al. Nasal carriage as a source of agr-defective Staphylococcus aureus bacteremia. J. Infect. Dis. 2012, 206, 1168–1177. [Google Scholar] [CrossRef]
- Loughman, J.A.; Fritz, S.A.; Storch, G.A.; Hunstad, D.A. Virulence Gene Expression in Human Community-Acquired Staphylococcus aureus Infection. J. Infect. Dis. 2009, 199, 294–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelinour, A.; Arvidson, S.; Bremell, T.; Ryden, C.; Tarkowski, A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 1993, 61, 3879–3885. [Google Scholar]
- Bunce, C.; Wheeler, L.; Reed, G.; Musser, J.; Barg, N. Murine model of cutaneous infection with gram-positive cocci. Infect. Immun. 1992, 60, 2636–2640. [Google Scholar] [PubMed]
- Cheung, A.L.; Eberhardt, K.J.; Chung, E.; Yeaman, M.R.; Sullam, P.M.; Ramos, M.; Bayer, A.S. Diminished virulence of a sar–/agr– mutant of Staphylococcus aureus in the rabbit model of endocarditis. J. Clin. Investig. 1994, 94, 1815–1822. [Google Scholar] [PubMed] [Green Version]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [PubMed] [Green Version]
- Khan, B.A.; Yeh, A.J.; Cheung, G.Y.; Otto, M. Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 2015, 24, 689–704. [Google Scholar] [CrossRef]
- Cancio, N.R.C.; Placeres, T.E.G. Method for Obtaining 2-bromo-5-(2-bromo-2-nitrovinyl)-furan. US20030130529A1, 10 July 2003. [Google Scholar]
- Cancio, N.R.C.; Rodriguez, S.S.; Fidalgo, L.M.; Hernandez, Y.L.; Alvarez, A.M.M.; Bourzac, J.F.I.; Manso, E.E.O. Pharmaceutical Compositions Containing Nitrovinylfuran Derivatives for the Treatment of Leishmaniosis and Trypanosomosis. EP1941877B1, 18 November 2009. [Google Scholar]
- Xiong, Y.-Q.; Van Wamel, W.; Nast, C.C.; Yeaman, M.R.; Cheung, A.L.; Bayer, A.S. Activation and transcriptional interaction between agr RNAII and RNAIII in Staphylococcus aureus in vitro and in an experimental endocarditis model. J. Infect. Dis. 2002, 186, 668–677. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Saavedra, M.J.; Simões, M. The activity of ferulic and gallic acids in biofilm prevention and control of pathogenic bacteria. Biofouling 2012, 28, 755–767. [Google Scholar] [CrossRef]
- Mohamadi, F.; Richards, N.G.J.; Guida, W.C.; Liskamp, R.; Lipton, M.; Caufield, C.; Chang, G.; Hendrickson, T.; Still, W.C. Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 1990, 11, 440–467. [Google Scholar] [CrossRef]
- Halgren, T. A Merck Molecular Force Field. II. MMFF94 van der Waals and Electrostatic Parameters for Intermolecular Interactions. J. Comput. Chem. 1996, 17, 520–552. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, A.E.; Guallar, V.; Berne, B.J.; Friesner, R. Importance of accurate charges in molecular docking: Quantum Mechanical/Molecular Mechanical (QM/MM) approach. J. Comput. Chem. 2005, 26, 915–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banks, J.L.; Beard, H.S.; Cao, Y.; Cho, A.E.; Damm, W.; Farid, R.; Felts, A.K.; Halgren, T.A.; Mainz, D.T.; Maple, J.R.; et al. Integrated Modeling Program, Applied Chemical Theory (IMPACT). J. Comput. Chem. 2005, 26, 1752–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Milena, S.-V. A modified microtiter—plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Borges, A.; Simões, L.C.; Saavedra, M.J.; Simões, M. The action of selected isothiocyanates on bacterial biofilm prevention and control. Int. Biodeterior. Biodegrad. 2014, 86, 25–33. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial Activity and Mode of Action of Ferulic and Gallic Acids Against Pathogenic Bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef]
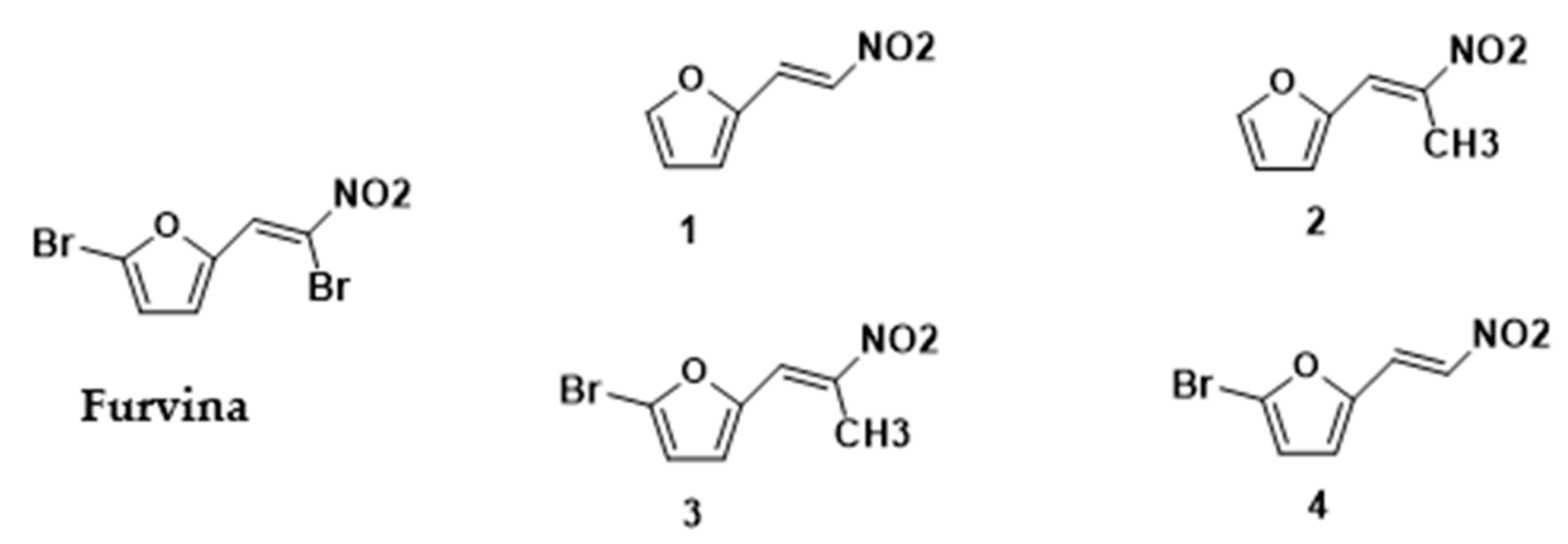
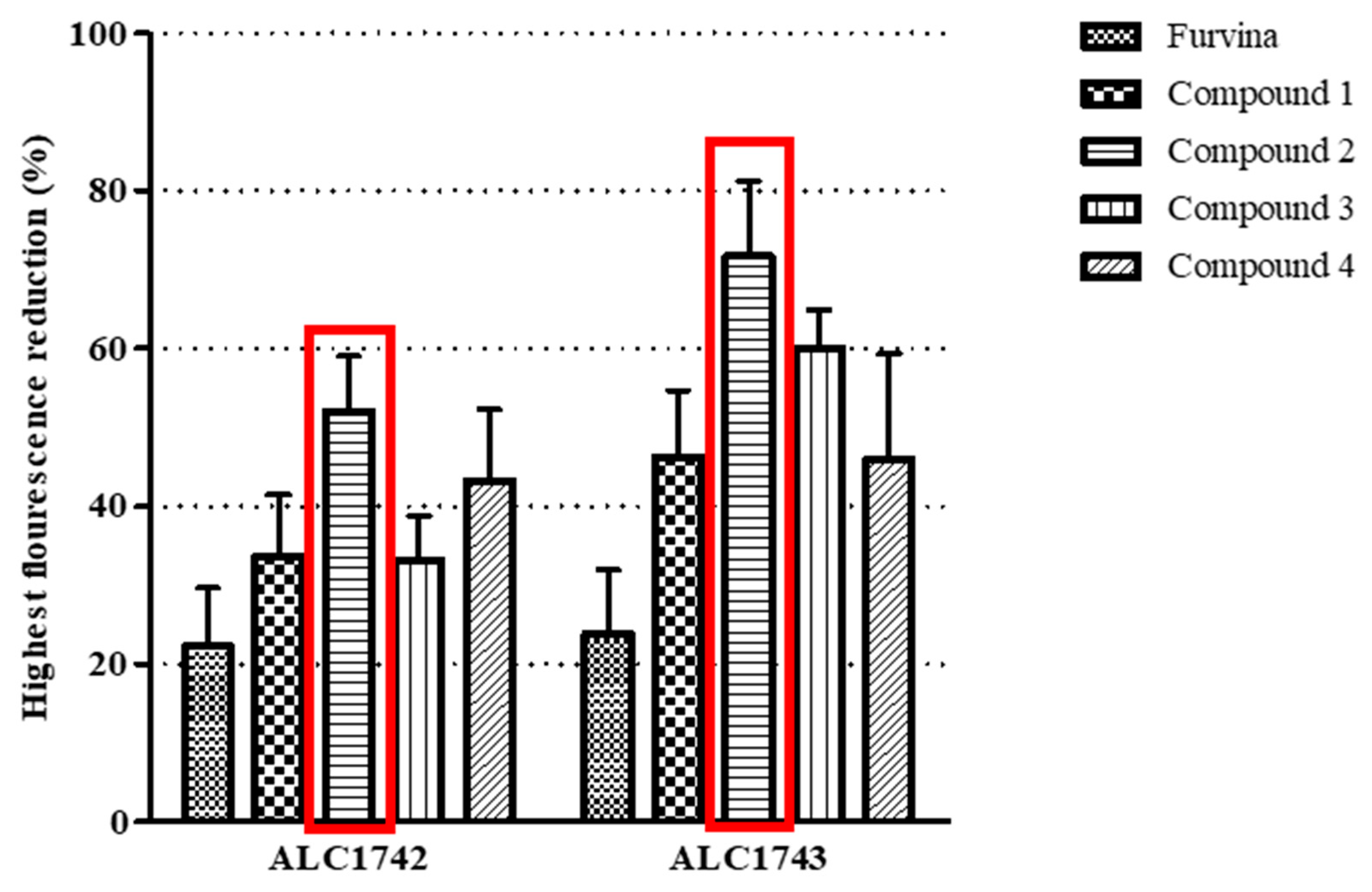
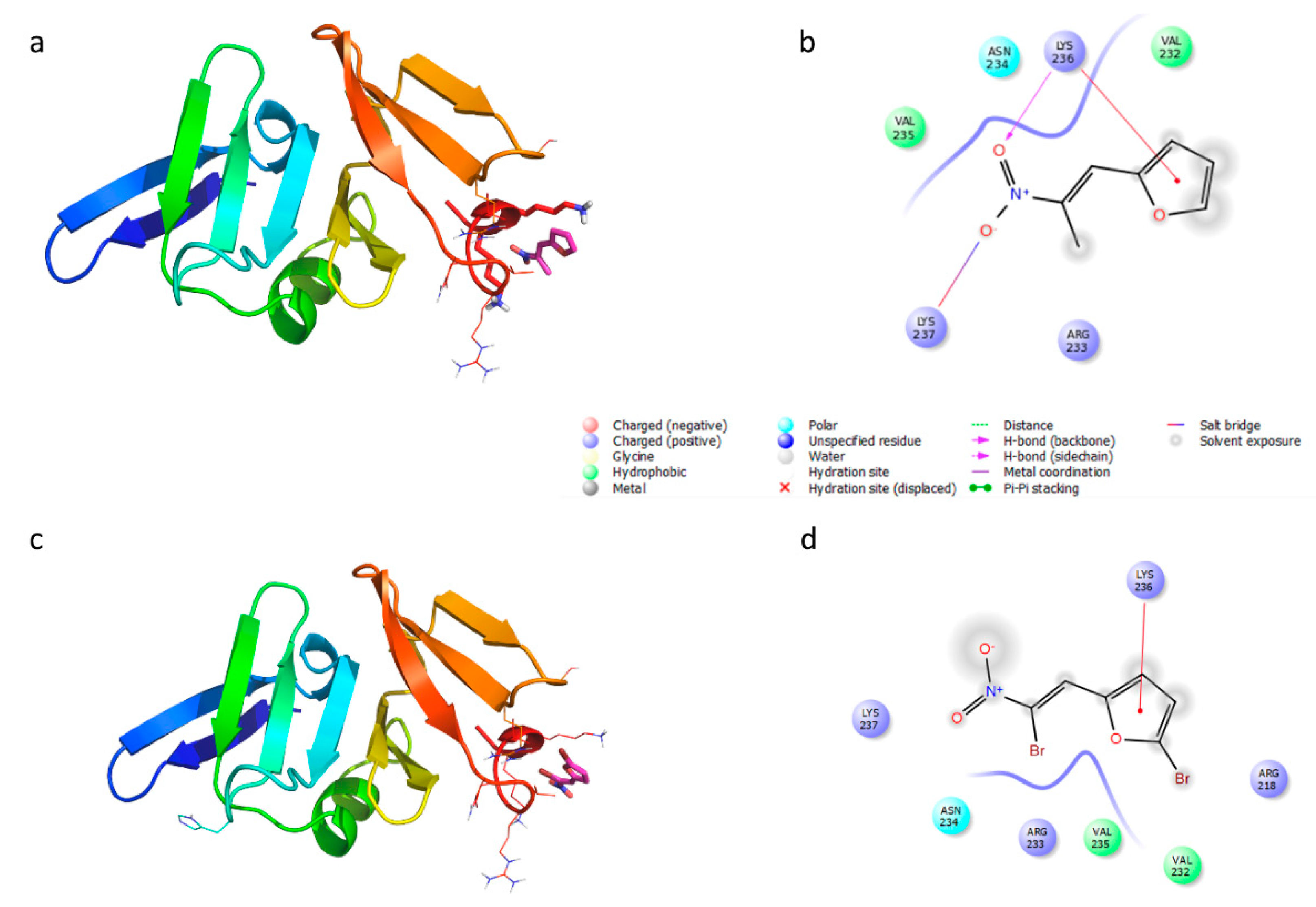
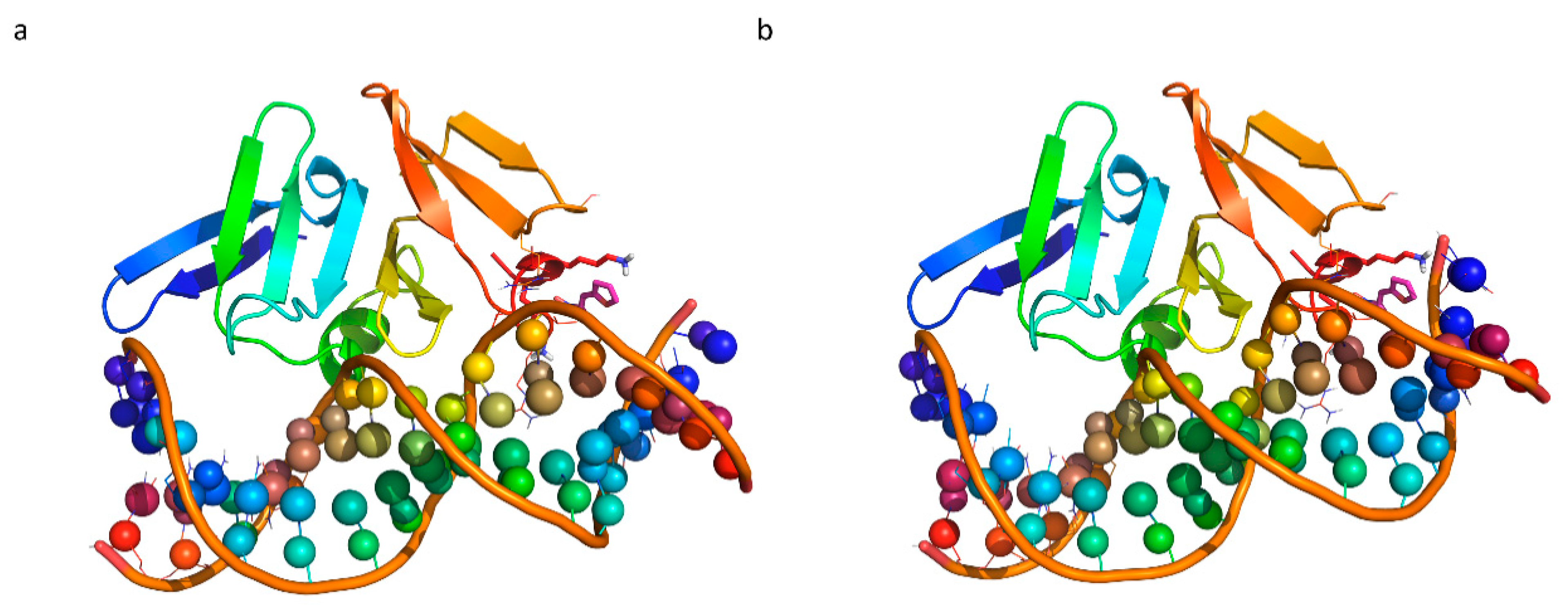
| Chemical Structure | Designation | MIC (µg/mL) | |
|---|---|---|---|
| S. aureus ALC1742 | S. aureus ALC1743 | ||
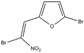 | Furvina | 100 | 100 |
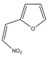 | Compound 1 | 500 | 400 |
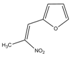 | Compound 2 | >1000 | >1000 |
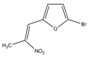 | Compound 3 | 400 | 400 |
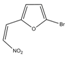 | Compound 4 | >1000 | >1000 |
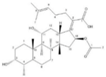 | Fusidic acid | 0.25 | 0.25 |
| Compound | Strain | Concentrations (μg/mL) | Biomass Reduction (%) | Metabolic Activity Reduction (%) | Intact Cells (%) |
|---|---|---|---|---|---|
| Furvina | ALC1742 | MIC (100) | 42.0 ± 8.0 | 75.1 ± 5.0 | 85.4 ± 8.1 |
| ½ MIC (50) | 37.5 ± 7.5 | 66.4 ± 8.0 | 86.1 ± 9.8 | ||
| ALC1743 | MIC (100) | 44.6 ± 1.7 | 78.6 ± 1.0 | 75.0 ± 18.6 | |
| ½ MIC (50) | 41.2 ± 5.4 | 77.8 ± 1.7 | 84.4 ± 14.6 | ||
| Compound 2 | ALC1742 | HCT of compound 2 (1000) | 19.0 ± 3.0 | 42.1 ±7.4 | 95.0 ± 3.0 |
| ½ HCT of compound 2 (500) | 17.5 ± 2.9 | 41.6 ± 8.5 | 93.6 ± 5.7 | ||
| ALC1743 | HCT of compound 2 (1000) | 17.3 ± 3.3 | 28.6 ± 8.6 | 94.0 ± 2.7 | |
| ½ HCT of compound 2 (500) | 21.5 ± 1.0 | 17.8 ± 4.1 | 93.7 ± 1.9 | ||
| Fusidic acid | ALC1742 | 10× MIC (2.5) | 19.97 ± 7.9 | 71.0 ± 5.9 | 91.5 ± 3.6 |
| ALC1743 | 10× MIC (2.5) | 22.7 ± 8.8 | 70.8 ± 6.6 | 87.8 ± 6.6 | |
| Fusidic acid + Compound 2 | ALC1742 | 10× MIC fusidic acid (2.5) + HCT of compound 2 (1000) | 33.7 ± 4.0 | 81.7 ± 5.8 | 66.9 ± 8.3 |
| ALC1743 | 10× MIC fusidic acid (2.5) + HCT of compound 2 (1000) | - * | 82.3 ± 4.0 | 81.1 ± 4.2 | |
| Fusidic acid + Furvina | ALC1742 | 10× MIC fusidic acid (2.5) + MIC (100) | 38.0 ± 14.4 | 72.6 ± 4.2 | 90.2 ± 4.7 |
| ALC1743 | 10× MIC fusidic acid (2.5) + MIC (100) | 28.9 ± 4.7 | 74.3 ± 4.3 | 85.6 ± 5.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, D.; Borges, A.; Ruiz, R.M.; Negrín, Z.R.; Distinto, S.; Borges, F.; Simões, M. 2-(2-Methyl-2-nitrovinyl)furan but Not Furvina Interfere with Staphylococcus aureus Agr Quorum-Sensing System and Potentiate the Action of Fusidic Acid against Biofilms. Int. J. Mol. Sci. 2021, 22, 613. https://doi.org/10.3390/ijms22020613
Oliveira D, Borges A, Ruiz RM, Negrín ZR, Distinto S, Borges F, Simões M. 2-(2-Methyl-2-nitrovinyl)furan but Not Furvina Interfere with Staphylococcus aureus Agr Quorum-Sensing System and Potentiate the Action of Fusidic Acid against Biofilms. International Journal of Molecular Sciences. 2021; 22(2):613. https://doi.org/10.3390/ijms22020613
Chicago/Turabian StyleOliveira, Diana, Anabela Borges, Reinaldo Molina Ruiz, Zenaida Rodríguez Negrín, Simona Distinto, Fernanda Borges, and Manuel Simões. 2021. "2-(2-Methyl-2-nitrovinyl)furan but Not Furvina Interfere with Staphylococcus aureus Agr Quorum-Sensing System and Potentiate the Action of Fusidic Acid against Biofilms" International Journal of Molecular Sciences 22, no. 2: 613. https://doi.org/10.3390/ijms22020613







