Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori
Abstract
:1. Introduction
2. Results
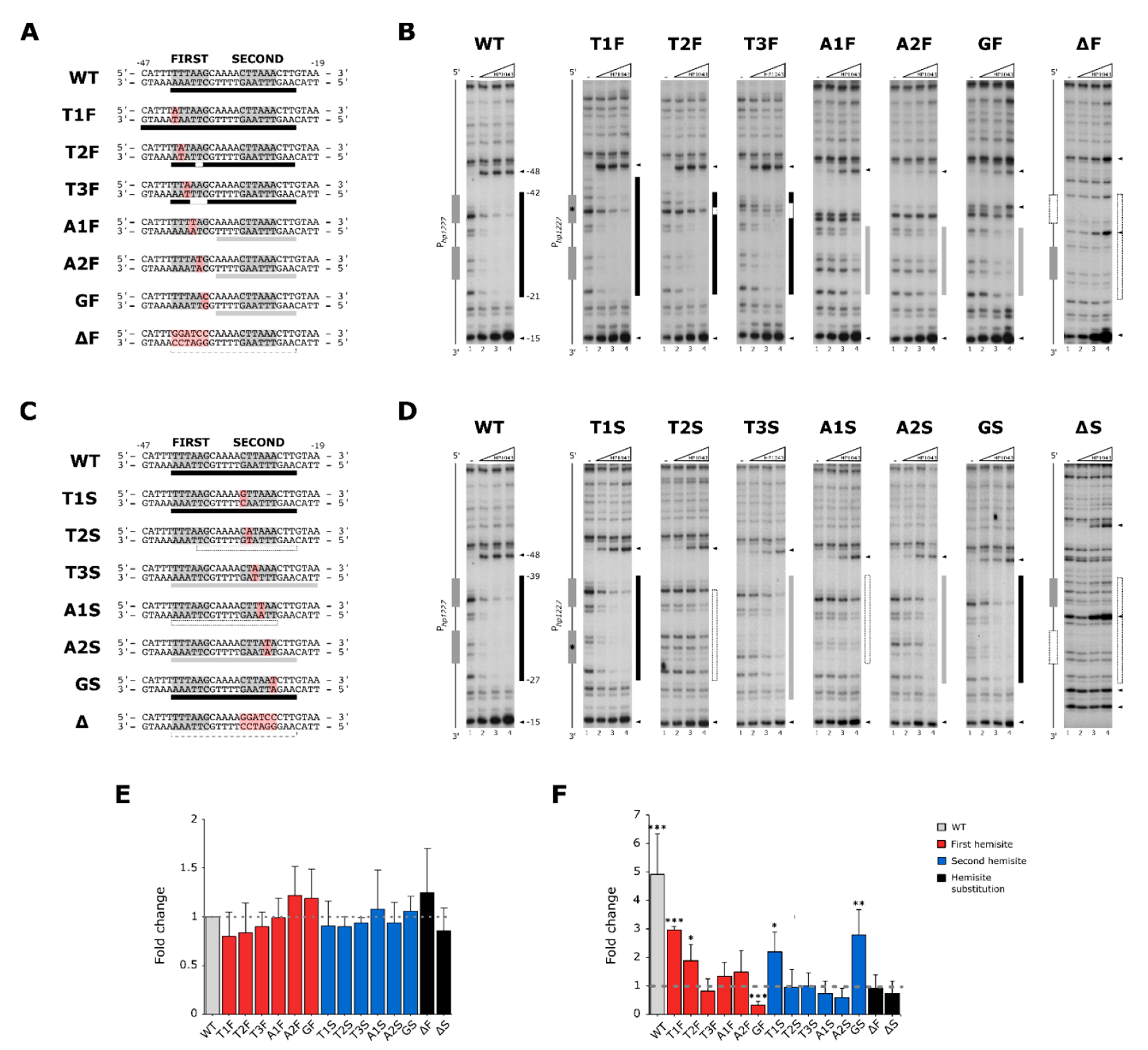
2.1. Molecular Characterization of HP1043 Consensus Binding Sequence
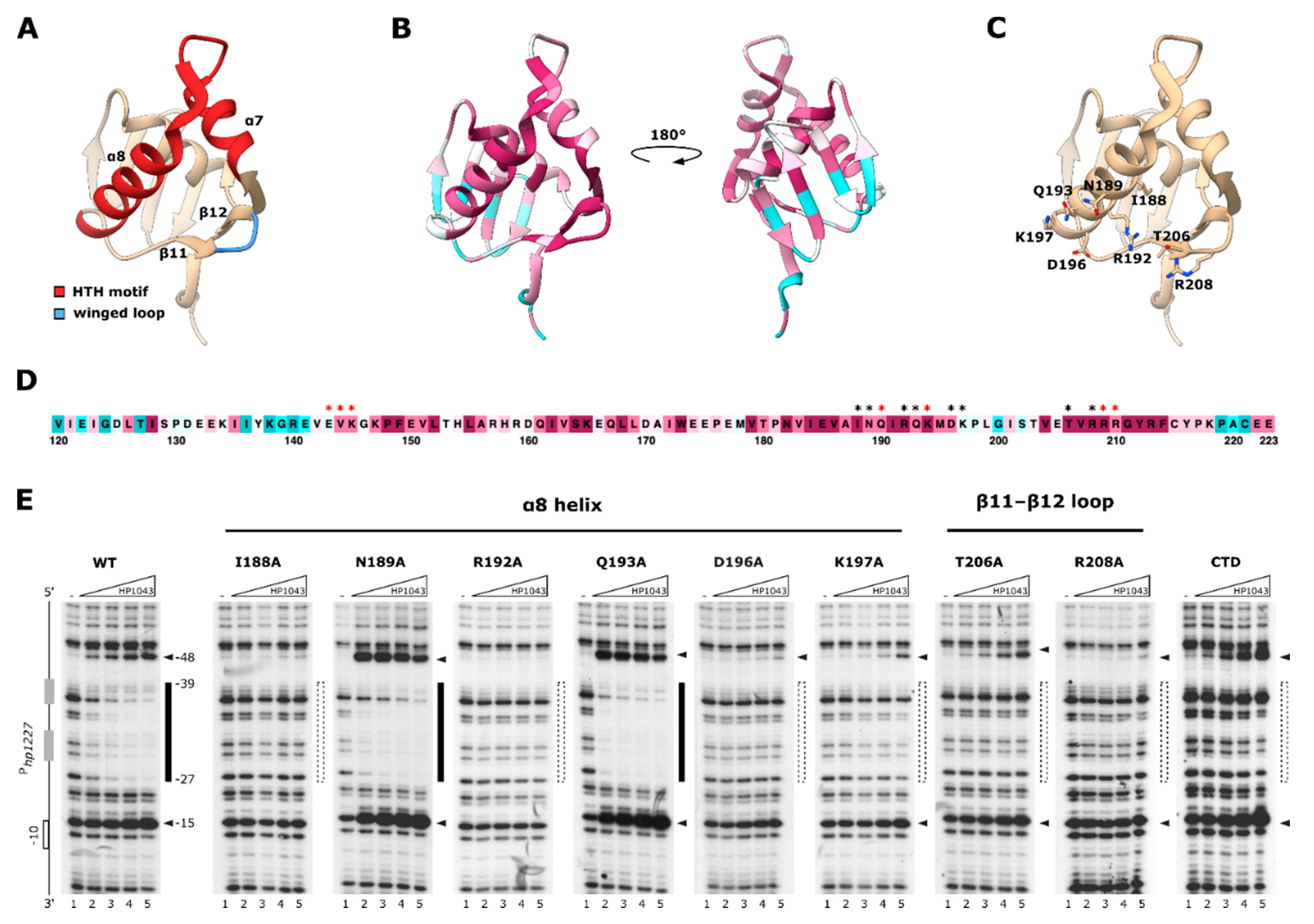
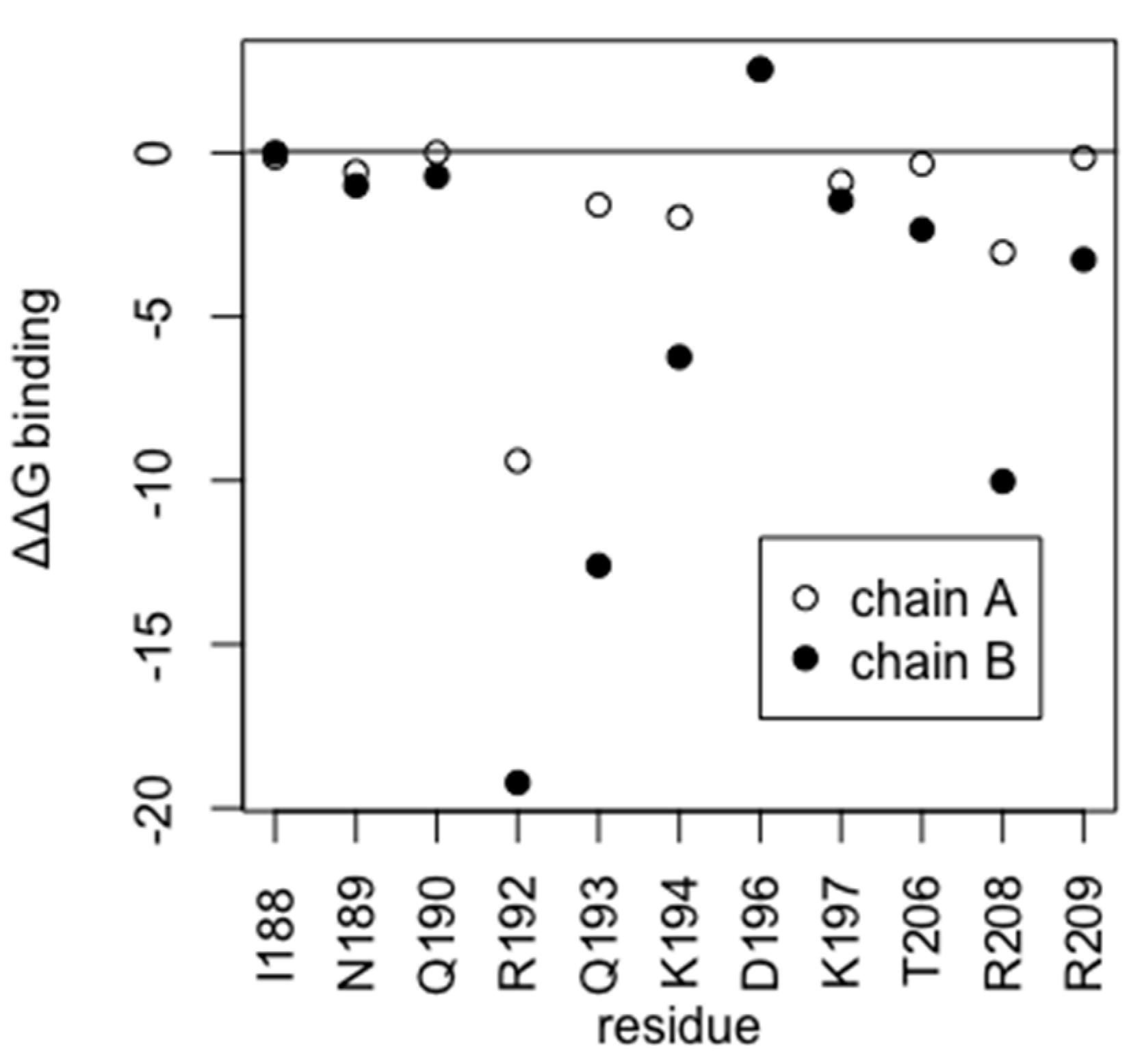
2.2. Identification of Amino Acid Residues Fundamental for Target DNA Recognition by HP1043
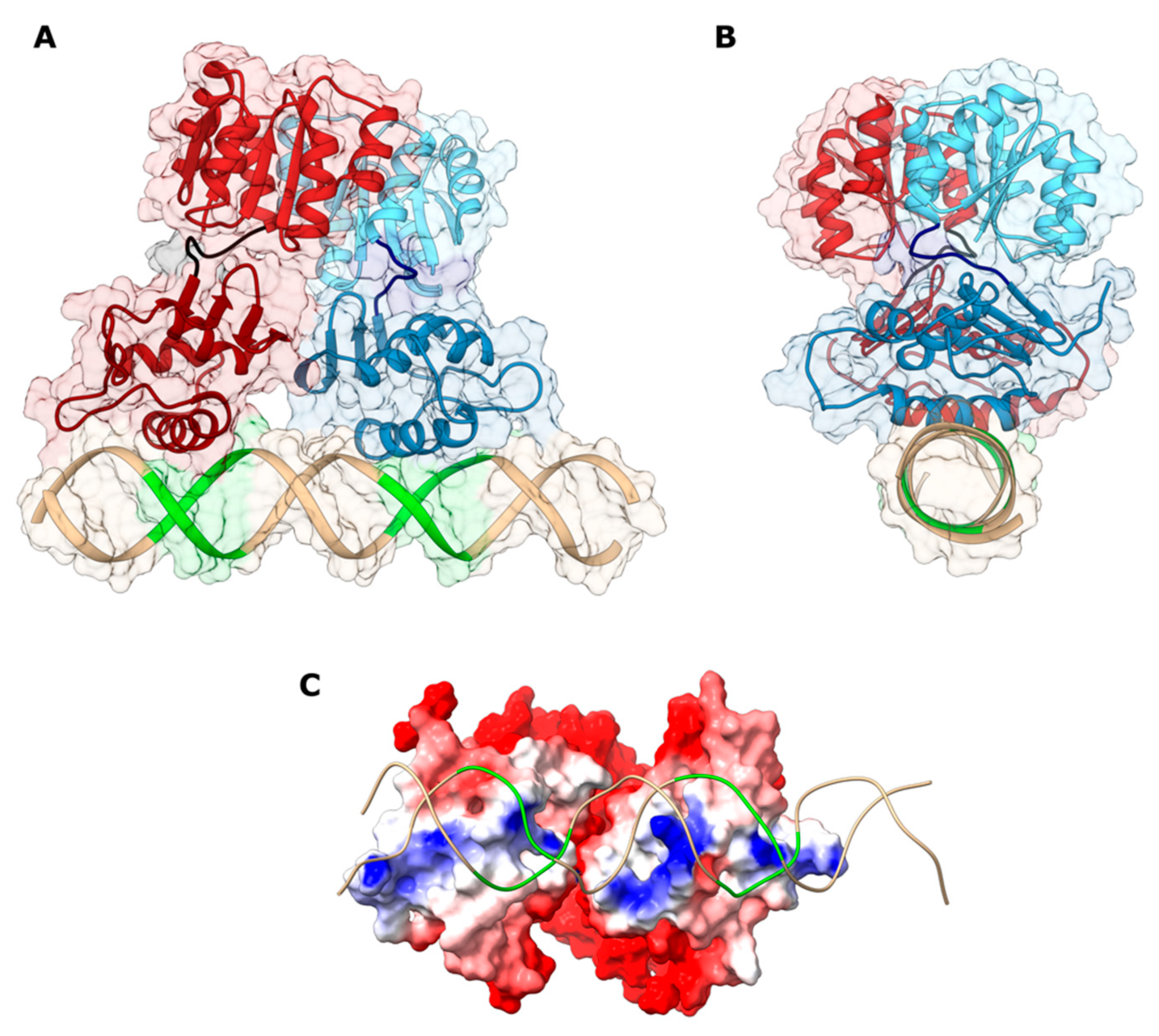
2.3. Structural Modeling of the HP1043_DBD-DNA Interaction
2.4. Validation of the HP1043-Php1227 Docking Model
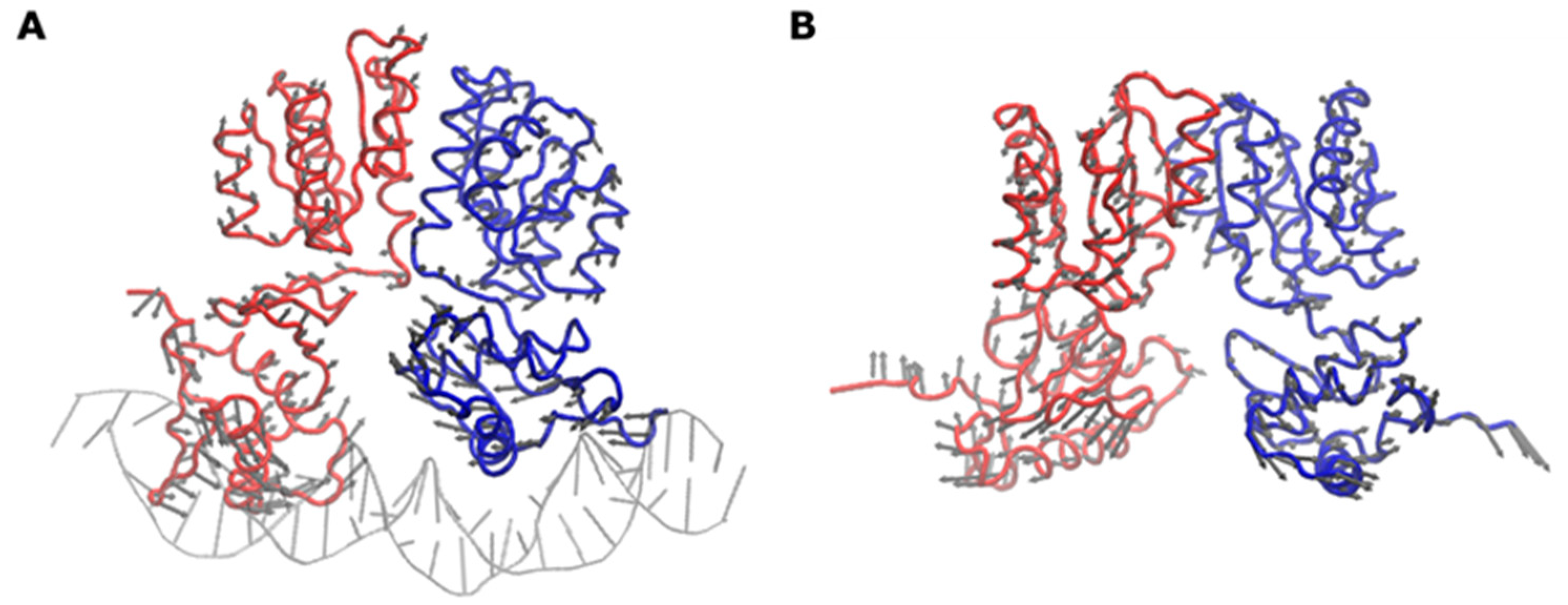
2.5. Conformational and Interaction Analysis of HP1043-DNA Full Model
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. DNA Manipulations
4.3. Overexpression and Purification of Recombinant His6-HP1043
4.4. DNase I Footprinting
4.5. Generation of Probes for DNase I Footprinting and Templates for In Vitro Transcription Experiments
4.6. In Vitro Transcription and cDNA Synthesis
4.7. qRT-PCR Analysis
4.8. Protein-DNA Docking and HP1043 Structure Reconstruction
4.9. Electrophoretic Mobility Shift Assays
4.10. Molecular Dynamics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Salama, N.R.; Hartung, M.L.; Muller, A. Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Micro. 2013, 11, 385–399. [Google Scholar] [CrossRef]
- Gisbert, J.P.; Calvet, X. Review article: Common misconceptions in the management of Helicobacter pylori-associated gastric MALT-lymphoma. Aliment. Pharmacol. Ther. 2011, 34, 1047–1062. [Google Scholar] [CrossRef] [PubMed]
- Scarlato, V.; Delany, I.; Spohn, G.; Beier, D. Regulation of transcription in Helicobacter pylori: Simple systems or complex circuits? Int. J. Med. Microbiol. 2001, 291, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Foynes, S.; Dorrell, N.; Ward, S.J.; Stabler, R.A.; McColm, A.A.; Rycroft, A.N.; Wren, B.W. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 2000, 68, 2016–2023. [Google Scholar] [CrossRef] [Green Version]
- Waidner, B.; Melchers, K.; Stähler, F.N.; Kist, M.; Bereswill, S. The Helicobacter pylori CrdRS two-component regulation system (HP1364/HP1365) is required for copper-mediated induction of the copper resistance determinant CrdA. J. Bacteriol. 2005, 187, 4683–4688. [Google Scholar] [CrossRef] [Green Version]
- Niehus, E.; Gressmann, H.; Ye, F.; Schlapbach, R.; Dehio, M.; Dehio, C.; Stack, A.; Meyer, T.F.; Suerbaum, S.; Josenhans, C. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 2004, 52, 947–961. [Google Scholar] [CrossRef]
- Pflock, M.; Finsterer, N.; Joseph, B.; Meyer, T.F.; Beier, D.; Mollenkopf, H. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 2006, 188, 3449–3462. [Google Scholar] [CrossRef] [Green Version]
- McDaniel, T.K.; DeWalt, K.C.; Salama, N.R.; Falkow, S. New Approaches for Validation of Lethal Phenotypes and Genetic Reversion in Helicobacter pylori. Helicobacter. 2001, 6, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Schär, J.; Sickmann, A.; Beier, D. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J. Bacteriol. 2005, 187, 3100–3109. [Google Scholar] [CrossRef] [Green Version]
- Delany, I.; Spohn, G.; Rappuoli, R.; Scarlato, V. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J. Bacteriol. 2002, 184, 4800–4810. [Google Scholar] [CrossRef] [Green Version]
- Müller, S.; Pflock, M.; Schär, J.; Kennard, S.; Beier, D. Regulation of expression of atypical orphan response regulators of Helicobacter pylori. Microbiol. Res. 2007, 162, 1–14. [Google Scholar] [CrossRef]
- Croxen, M.A.; Ernst, P.B.; Hoffman, P.S. Antisense RNA Modulation of Alkyl Hydroperoxide Reductase Levels in Helicobacter pylori Correlates with Organic Peroxide Toxicity but Not Infectivity. J. Bacteriol. 2007, 189, 3359–3368. [Google Scholar] [CrossRef] [Green Version]
- Pelliciari, S.; Pinatel, E.; Vannini, A.; Peano, C.; Puccio, S.; De Bellis, G.; Danielli, A.; Scarlato, V.; Roncarati, D. Insight into the essential role of the Helicobacter pylori HP1043 orphan response regulator: Genome-wide identification and characterization of the DNA-binding sites. Sci. Rep. 2017, 7, 41063. [Google Scholar] [CrossRef] [PubMed]
- Browning, D.F.; Busby, S.J.W. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016, 14, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Hong, E.; Jung, J.-W.; Shin, J.; Kim, J.H.; Jeon, Y.-H.; Yamazaki, T.; Lee, W. Letter to the Editor: Backbone 1 H, 13 C and 15 N resonance assignments of the response regulator HP1043 from Helicobactor pylori. J. Biomol. NMR 2004, 28, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Hyang, M.L.; Ko, H.; Kim, D.U.; Jeon, B.Y.; Jung, J.; Shin, J.; Lee, S.A.; Kim, Y.; Young, H.J.; et al. Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J. Biol. Chem. 2007, 282, 20667–20675. [Google Scholar] [CrossRef] [Green Version]
- Jeong, K.W.; Ko, H.; Lee, S.A.; Hong, E.; Ko, S.; Cho, H.S.; Lee, W.; Kim, Y. Backbone dynamics of an atypical orphan response regulator protein, Helicobacter pylori 1043. Mol. Cells 2013, 35, 158–165. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, M.; Van Dijk, A.D.J.; Hsu, V.; Rolf, B.; Bonvin, A.M.J.J. Information-driven protein-DNA docking using HADDOCK: It is a matter of flexibility. Nucleic Acids Res. 2006, 34, 3317–3325. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.G.; Sola, M.; Gomis-Rüth, F.X.; Coll, M. Tandem DNA Recognition by PhoB, a Two-Component Signal Transduction Transcriptional Activator. Structure 2002, 10, 701–713. [Google Scholar] [CrossRef]
- Li, Y.-C.; Chang, C.; Chang, C.-F.; Cheng, Y.-H.; Fang, P.-J.; Yu, T.; Chen, S.-C.; Li, Y.-C.; Hsiao, C.-D.; Huang, T. Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element. Nucleic Acids Res. 2014, 42, 8777–8788. [Google Scholar] [CrossRef] [Green Version]
- Lou, Y.-C.; Weng, T.-H.; Li, Y.-C.; Kao, Y.-F.; Lin, W.-F.; Peng, H.-L.; Chou, S.-H.; Hsiao, C.-D.; Chen, C. Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA. Nat. Commun. 2015, 6, 8838. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors 1 1Edited by M. Gottesman. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Tanaka, I. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol. Microbiol. 1997, 24, 665–667. [Google Scholar] [CrossRef]
- Rhee, J.E.; Sheng, W.; Morgan, L.K.; Nolet, R.; Liao, X.; Kenney, L.J. Amino Acids Important for DNA Recognition by the Response Regulator OmpR. J. Biol. Chem. 2008, 283, 8664–8677. [Google Scholar] [CrossRef] [Green Version]
- Walthers, D.; Tran, V.K.; Kenney, L.J. Interdomain Linkers of Homologous Response Regulators Determine Their Mechanism of Action. J. Bacteriol. 2003, 185, 317–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, J.J.; Moughon, S.; Wang, C.; Schueler-Furman, O.; Kuhlman, B.; Rohl, C.A.; Baker, D. Protein–Protein Docking with Simultaneous Optimization of Rigid-body Displacement and Side-chain Conformations. J. Mol. Biol. 2003, 331, 281–299. [Google Scholar] [CrossRef]
- Marze, N.A.; Roy Burman, S.S.; Sheffler, W.; Gray, J.J. Efficient flexible backbone protein–protein docking for challenging targets. Bioinformatics 2018, 34, 3461–3469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Tsui, V.; Case, D.A. Molecular Dynamics Simulations of Nucleic Acids with a Generalized Born Solvation Model. J. Am. Chem. Soc. 2000, 122, 2489–2498. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Gonzalez, A.; Fillat, M.F.; Lanas, Á. Transcriptional regulators: Valuable targets for novel antibacterial strategies. Future Med. Chem. 2018, 10, 541–560. [Google Scholar] [CrossRef]
- González, A.; Salillas, S.; Velázquez-Campoy, A.; Espinosa Angarica, V.; Fillat, M.F.; Sancho, J.; Lanas, Á. Identifying potential novel drugs against Helicobacter pylori by targeting the essential response regulator HsrA. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Casado, J.; Chueca, E.; Salillas, S.; Velázquez-Campoy, A.; Espinosa Angarica, V.; Bénejat, L.; Guignard, J.; Giese, A.; Sancho, J.; et al. Repurposing Dihydropyridines for Treatment of Helicobacter pylori Infection. Pharmaceutics 2019, 11, 681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York, NY, USA, 1989; pp. 1–1546. [Google Scholar]
- Liu, S.-T.; Hong, G.-F. Three-Minute G+A Specific Reaction for DNA Sequencing. Anal. Biochem. 1998, 255, 158–159. [Google Scholar] [CrossRef] [PubMed]
- Winson, M.K.; Swift, S.; Hill, P.J.; Sims, C.M.; Griesmayr, G.; Bycroft, B.W.; Williams, P.; Stewart, G.S.A. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn 5 constructs. FEMS Microbiol. Lett. 1998, 163, 193–202. [Google Scholar] [CrossRef] [Green Version]
- de Vries, S.J.; van Dijk, A.D.J.; Krzeminski, M.; van Dijk, M.; Thureau, A.; Hsu, V.; Wassenaar, T.; Bonvin, A.M.J.J. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins Struct. Funct. Bioinforma 2007, 69, 726–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agriesti, F.; Roncarati, D.; Musiani, F.; Del Campo, C.; Iurlaro, M.; Sparla, F.; Ciurli, S.; Danielli, A.; Scarlato, V. FeON-FeOFF: The Helicobacter pylori Fur regulator commutates iron-responsive transcription by discriminative readout of opposed DNA grooves. Nucleic Acids Res. 2014, 42, 3138–3151. [Google Scholar] [CrossRef] [Green Version]
- Mazzei, L.; Dobrovolska, O.; Musiani, F.; Zambelli, B.; Ciurli, S. On the interaction of Helicobacter pylori NikR, a Ni(II)-responsive transcription factor, with the urease operator: In solution and in silico studies. J. Biol. Inorg. Chem. 2015, 20, 1021–1037. [Google Scholar] [CrossRef]
- van Dijk, M.; Bonvin, A.M.J.J. 3D-DART: A DNA structure modelling server. Nucleic Acids Res. 2009, 37, W235–W239. [Google Scholar] [CrossRef]
- Musiani, F.; Ciurli, S.; Dikiy, A. Interaction of Selenoprotein W with 14-3-3 Proteins: A Computational Approach. J. Proteome Res. 2011, 10, 968–976. [Google Scholar] [CrossRef]
- Shen, M.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [Green Version]
- Honig, B.; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomb, J.-F.; White, O.; Kerlavage, A.R.; Clayton, R.A.; Sutton, G.G.; Fleischmann, R.D.; Ketchum, K.A.; Klenk, H.P.; Gill, S.; Dougherty, B.A.; et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997, 388, 539–547. [Google Scholar] [CrossRef] [Green Version]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [Green Version]
- Cheatham, T.E.; Case, D.A. Twenty-five years of nucleic acid simulations. Biopolymers 2013. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Onufriev, A.; Bashford, D.; Case, D.A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins Struct. Funct. Bioinforma 2004, 55, 383–394. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zannoni, A.; Pelliciari, S.; Musiani, F.; Chiappori, F.; Roncarati, D.; Scarlato, V. Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori. Int. J. Mol. Sci. 2021, 22, 7848. https://doi.org/10.3390/ijms22157848
Zannoni A, Pelliciari S, Musiani F, Chiappori F, Roncarati D, Scarlato V. Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori. International Journal of Molecular Sciences. 2021; 22(15):7848. https://doi.org/10.3390/ijms22157848
Chicago/Turabian StyleZannoni, Annamaria, Simone Pelliciari, Francesco Musiani, Federica Chiappori, Davide Roncarati, and Vincenzo Scarlato. 2021. "Definition of the Binding Architecture to a Target Promoter of HP1043, the Essential Master Regulator of Helicobacter pylori" International Journal of Molecular Sciences 22, no. 15: 7848. https://doi.org/10.3390/ijms22157848







