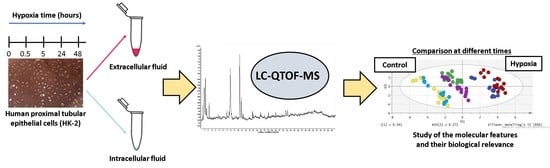
Exploratory Metabolomic Analysis Based on Reversed-Phase Liquid Chromatography–Mass Spectrometry to Study an In Vitro Model of Hypoxia-Induced Metabolic Alterations in HK-2 Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Untargeted Metabolomics Analysis of an In Vitro Model of Hypoxia in HK-2 Cells
2.1.1. Metabolomic Analysis at Hypoxia Times of 0.5 and 5 h (Short-Term Hypoxia)
2.1.2. Metabolomic Analysis at Hypoxia Times of 24 and 48 h (Long-Term Hypoxia)
2.2. Evaluation of the Metabolic Effects Observed under Short and Long Times of Hypoxia
2.3. Biological Interpretation
3. Materials and Methods
3.1. Reagents and Solvents
3.2. HK-2 Cell Line Culture
3.3. Protein Isolation and Western-Blotting
3.4. Optimized Sample Preparation Protocol
3.5. Liquid Chromatography–Mass Spectrometry Analysis
3.6. Metabolomic Sequence
3.7. Data Treatment and Analysis
3.8. Identification of Metabolites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pouyssegur, J.; Lopez-Barneo, J. Hypoxia in health and disease. Mol. Asp. Med. 2016, 47–48, 1–2. [Google Scholar] [CrossRef]
- Mylonis, I.; Simos, G.; Paraskeva, E. Hypoxia-inducible factors and the regulation of lipid metabolism. Cells 2019, 8, 214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semenza, G.L. Hypoxia-inducible factors in physiology and medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirakawa, Y.; Tanaka, T.; Nangaku, M. Renal hypoxia in CKD; pathophysiology and detecting methods. Front. Physiol. 2017, 8, 99. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.S.; Chiu, W.T.; Hsu, P.L.; Lin, S.C.; Peng, I.C.; Wang, C.Y.; Tsai, S.J. Pathophysiological implications of hypoxia in human diseases. J. Biomed. Sci. 2020, 27, 63. [Google Scholar] [CrossRef] [PubMed]
- Vinovskia, C.; Li, L.P.; Prasad, P.; Tommerdahl, K.; Pyle, L.; Nelson, R.G.; Pavkov, M.E.; van Raalte, D.; Rewers, M.; Pragnell, M.; et al. Relative hypoxia and early diabetic kidney disease in type 1 diabetes. Diabetes 2020, 69, 2700–2708. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Xu, G. Current and future perspectives of functional metabolomics in disease studies-a review. Anal. Chim. Acta 2018, 1037, 41–54. [Google Scholar] [CrossRef]
- Hounoum, B.M.; Blasco, H.; Emond, P.; Mavel, S. Liquidchromatography-high-resolution mass spectrometry-based cell metabolomics: Experimental design, recommendations, and applications. TrAC Trend. Anal. Chem. 2016, 75, 118–128. [Google Scholar] [CrossRef]
- Cuperlovic-Culf, M.; Barnett, D.A.; Culf, A.S.; Chute, I. Cell culture metabolomics: Applications and future directions. Drug Discov. Today 2010, 15, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.L.; Kuo, T.C.; Ho, T.J.; Harn, Y.C.; Wang, S.Y.; Fu, W.M.; Kuo, C.H.; Tseng, Y.F. Metabolomic dynamic analysis of hypoxia in MDAMB-231 and the comparison with inferred metabolites from transcriptomics data. Cancers 2013, 5, 491–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacchi, P.S.; Bloise, A.C.; Bustos, S.O.; Zimmermann, L.; Chammas, R.; Rabbani, S.R. Metabolism under hypoxia in Tm1 murine melanoma cells is affected by the presence of galectin-3, a metabolomics approach. SpringerPlus 2014, 3, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valli, A.; Rodriguez, M.; Moutsianas, L.; Fischer, R.; Fedele, V.; Huang, H.L.; Van Stiphout, R.; Jones, D.; Mccarthy, M.; Vinaxia, M. Hypoxia induces a lipogenic cancer cell phenotype via HIF1 alpha-dependent and -independent pathways. Oncotarget 2015, 6, 1920–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armitage, E.G.; Kotze, H.L.; Allwood, J.W.; Dunn, W.B.; Goodacre, R.; Williams, K.J. Metabolic profiling reveals potential metabolic markers associated with Hypoxia Inducible Factor-mediated signalling in hypoxic cancer cells. Sci. Rep. 2015, 5, 15649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Contreras, M.; Tamayo-Garcia, A.; Pappan, K.L.; Michelotti, G.A.; Stabler, C.L.; Ricordi, C.; Buchwald, P. Metabolomics study of the effects of inflammation, hypoxia, and high glucose on isolated human pancreatic islets. J. Proteome Res. 2017, 16, 2294–2306. [Google Scholar] [CrossRef]
- Bernardo-Bermejo, S.; Sánchez-López, E.; Castro-Puyana, M.; Benito, S.; Lucio-Cazaña, F.J.; Marina, M.L. An untargeted metabolomic strategy based on liquid chromatography-mass spectrometry to study high glucose-induced changes in HK-2 cells. J. Chromatogr. A. 2019, 1596, 124–133. [Google Scholar] [CrossRef]
- Bernardo-Bermejo, S.; Sánchez-López, E.; Castro-Puyana, M.; Benito, S.; Lucio-Cazaña, F.J.; Marina, M.L. A non-targeted capillary electrophoresis-mass spectrometry strategy to study metabolic differences in an in vitro model of model high-glucose induced changes in human proximal tubular Hk-2 cells. Molecules 2020, 25, 512. [Google Scholar] [CrossRef] [Green Version]
- Valdes, A.; Lucio-Cazaña, F.J.; Castro-Puyana, M.; Garcia-Pastor, C.; Fiehn, O.; Marina, M.L. Comprehensive metabolomic study of the response of Hk-2 cells to hyperglicemic hypoxic diabetic-like milieu. Sci. Rep. 2021, 11, 5058. [Google Scholar] [CrossRef]
- Fritsche-Guenther, R.; Bauer, A.; Gloaguen, Y.; Lorenz, M.; Kirwan, J.A. Modified protocol of harvesting, extraction, and normalization approaches for gas chromatography mass spectrometry-based metabolomics analysis of adherent cells grown under high fetal calf serum conditions. Metabolites 2019, 10, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Shi, H.; Zhang, R.; Wei, Z.; Bi, K.S.; Chen, X.H. Simultaneous Determination of Five Specific and Sensitive Nephrotoxicity Biomarkers in Serum and Urine Samples of Four Drug-Induced Kidney Injury Models. J. Chromatogr. Sci. 2017, 55, 60–68. [Google Scholar] [CrossRef]
- Stumvoll, M.; Perriello, G.; Meyer, C.; Gerich, J. Role of glutamine in human carbohydrate metabolism in kidney and other tissues. Kidney Int. 1999, 55, 778–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, T.; Hirakawa, Y.; Nangaku, M. The role of oxidative stress and hypoxia in renal disease. Kidney Res. Clin. Pract. 2019, 38, 414–426. [Google Scholar] [CrossRef] [Green Version]
- Eckardt, K.U.; Bernhardt, W.M.; Weidemann, A.; Warnecke, C.; Rosenberger, C.; Wiesener, M.S.; Willam, C. Role of hypoxia in the pathogenesis of renal disease. Kidney Int. Suppl. 2005, 68, S46–S51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.; Zampino, M.; Moaddel, R.; Chen, T.K.; Tian, Q.; Ferrucci, L.; Semba, R.D. Plasma metabolites associated with chronic kidney disease and renal function in adults from the Baltimore Longitudinal Study of Aging. Metabolomics 2021, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.G.; Diamond, M.; Waddell, J.; McKenna, M.C. Effect of acetyl-l-carnitine used for protection of neonatal hypoxic-ischemic brain injury on acute kidney changes in male and female rats. Neurochem. Res. 2019, 44, 2405–2412. [Google Scholar] [CrossRef]
- Bhargava, P.; Schnellmann, R.G. Mitochondrial energetics in the kidney. Nat. Rev. Nephrol. 2017, 13, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Aguer, C.; McCoin, C.S.; Knotts, T.A.; Thrush, A.B.; Ono-Moore, K.; McPherson, R.; Dent, R.; Hwang, D.H.; Adams, S.H.; Harper, M.E. Acylcarnitines: Potential implications for skeletal muscle insulin resistance. FASEB J. 2015, 29, 336–345. [Google Scholar] [CrossRef] [Green Version]
- Rebouche, C.J.; Lombard, K.A.; Chenard, C.A. Renal adaptation to dietary carnitine in humans. Am. J. Clin. Nutr. 1993, 58, 660–665. [Google Scholar] [CrossRef]
- Bowman, B.B.; McCormick, D.B. Pyridoxine uptake by rat renal proximal tubular cells. J. Nutr. 1989, 119, 745–749. [Google Scholar] [CrossRef]
- Zhang, Z.M.; McCormick, D.B. Uptake of N-(4′-pyridoxyl)amines and release of amines by renal cells: A model for transporter-enhanced delivery of bioactive compounds. Proc. Natl. Acad. Sci. USA 1991, 88, 10407–10410. [Google Scholar] [CrossRef] [Green Version]
- Said, H.M.; Ortiz, A.; Vaziri, N.D. Mechanism and regulation of vitamin B(6)uptake by renal tubular epithelia: Studies with cultured OK cells. Am. J. Physiol. Renal Physiol. 2002, 282, 465–471. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Franco, M. Pyridoxine 5′-phosphate synthase: De novo synthesis of vitamin B6 and beyond. Biochim. Biophys. Acta 2003, 1647, 92–97. [Google Scholar] [CrossRef]
- Abosamak, N.E.R.; Gupta, V. Vitamin B6 (Pyridoxine). In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Stover, P.J.; Field, M.S. Vitamin B-6. Adv. Nutr. 2015, 6, 132–133. [Google Scholar] [CrossRef] [PubMed]
- Weljie, A.M.; Jirik, F.R. Hypoxia-induced metabolic shifts in cancer cells: Moving beyond the Warburg effect. Int. J. Biochem. Cell Biol. 2011, 43, 981–989. [Google Scholar] [CrossRef]
- Willetts, P.; Crossley, D.N.; Ramsden, D.B.; Hoffenberg, R. The role of thyronine in thyroid hormone metabolism. J. Clin. Endocrinol. Metab. 1979, 49, 658–660. [Google Scholar] [CrossRef]
- Leonard, J.L.; Ekenbarger, D.M.; Frank, S.J.; Farwell, A.P.; Koehrle, J. Localization of type I iodothyronine 5′-deiodinase to the basolateral plasma membrane in renal cortical epithelial cells. J. Biol. Chem. 1991, 266, 11262–11269. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, R.; Shen, G.; Feng, J. Metabolic response in rabbit urine to occurrence and relief of unilateral ureteral obstruction. J. Proteome Res. 2018, 17, 3184–3194. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, M.; Bo, L.; Li, S.; Hu, L.; Zhao, X.; Sun, C. Metabolomic analysis of the toxic effect of chronic exposure of cadmium on rat urine. Environ. Sci. Pollut. Res. Int. 2018, 25, 3765–3774. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Tang, D.D.; Chen, H.; Mao, J.R.; Bai, X.; Cheng, X.H.; Xiao, X.Y. Urinary metabolomics and biomarkers of aristolochic acid nephrotoxicity by UPLC-QTOF/HDMS. Bioanalysis 2015, 7, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Hine, D.G.; West-Dull, A.; Lynn, T.B. Gas-chromatographic method of analysis for urinary organic acids. I. Retention indices of 155 metabolically important compounds. Clin. Chem. 1980, 26, 1839–1846. [Google Scholar] [CrossRef]
- Kumar Pasupulati, A.; Chitra, P.S.; Reddy, G.B. Advanced glycation end products mediated cellular and molecular events in the pathology of diabetic nephropathy. Biomol. Concepts 2016, 7, 293–309. [Google Scholar] [CrossRef] [Green Version]
- Jørgensen, K.E.; Kragh-Hansen, U.; Sheikh, M.I. Transport of leucine, isoleucine and valine by luminal membrane vesicles from rabbit proximal tubule. J. Physiol. 1990, 422, 41–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachhawat, A.K.; Yadav, S. The glutathione cycle: Glutathione metabolism beyond the gamma-glutamyl cycle. IUBMB Life 2018, 70, 585–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Pol, A.; Gil, A.; Silljé, H.H.W.; Tromp, J.; Ovchinnikova, E.S.; Vreeswijk-Baudoin, I.; Hoes, M.; Domian, I.J.; van de Sluis, B.; van Deursen, J.M.; et al. Accumulation of 5-oxoproline in myocardial dysfunction and the protective effects of OPLAH. Sci. Transl. Med. 2017, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pederzolli, C.D.; Mescka, C.P.; Zandoná, B.R.; de Moura Coelho, D.; Sgaravatti, A.M.; Sgarbi, M.B.; de Souza Wyse, A.T.; Duval Wannmacher, C.M.; Wajner, M.; Vargas, C.R.; et al. Acute administration of 5-oxoprolineinduces oxidative damage to lipids and proteins and impairs antioxidant defenses in cerebral cortex and cerebellum of young rats. Metab. Brain Dis. 2010, 25, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Farías, J.G.; Herrera, E.A.; Carrasco-Pozo, C.; Sotomayor-Zárate, R.; Cruz, G.; Morales, P.; Castillo, R.L. Pharmacological models and approaches for pathophysiological conditions associated with hypoxia and oxidative stress. Pharmacol. Ther. 2016, 158, 1–23. [Google Scholar] [CrossRef]
- Temellini, A.; Mogavero, S.; Giulianotti, P.C.; Pietrabissa, A.; Mosca, F.; Pacifici, G.M. Conjugation of benzoic acid with glycine in human liver and kidney: A study on the interindividual variability. Xenobiotica 1993, 23, 1427–1433. [Google Scholar] [CrossRef]
- Ma, C.; Bi, K.; Zhang, M.; Su, D.; Fan, X.; Ji, W.; Wang, C.; Chen, X. Toxicology effects of morning glory seed in rat: A metabonomic method for profiling of urine metabolic changes. J. Ethnopharmacol. 2010, 130, 134–142. [Google Scholar] [CrossRef]
- Lees, H.J.; Swann, J.R.; Wilson, I.D.; Nicholson, J.K.; Holmes, E. Hippurate: The natural history of a mammalian-microbial cometabolite. J. Proteome Res. 2013, 12, 1527–1546. [Google Scholar] [CrossRef]
- Itoh, Y.; Ezawa, A.; Kikuchi, K.; Tsuruta, Y.; Niwa, T. Protein-bound uremic toxins in hemodialysis patients measured by liquid chromatography/tandem mass spectrometry and their effects on endothelial ROS production. Anal. Bioanal. Chem. 2012, 403, 1841–1850. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wei, R.; Wang, Y.; Su, T.; Li, P.; Chen, X. The uremic toxin hippurate promotes endothelial dysfunction via the activation of Drp1-mediated mitochondrial fission. Redox Biol. 2018, 16, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Gil de la Fuente, A.; Godzien, J.; Fernández López, V.; Rupérez, F.J.; Barbas, C.; Otero, A. Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef] [PubMed]




| PLS-DA Models | R2X | R2Y | Q2 | CV-ANOVA | |
|---|---|---|---|---|---|
| Intracellular fluid | |||||
| Ht0.5h vs. Ct0.5h | 0.620 | 0.801 | 0.732 | F(17.1) | p(7.3 × 10−7) |
| Ht5h vs. Ct5h | 0.794 | 0.982 | 0.968 | F(102.8) | p(1.9 × 10−15) |
| Extracellular fluid | |||||
| Ht0.5h vs. Ct0.5h | 0.606 | 0.998 | 0.986 | F(186.7) | p(1.2 × 10−16) |
| Ht5h vs. Ct5h | 0.652 | 0.996 | 0.978 | F(117.0) | p(1.5 × 10−15) |
| # | RT (min) | Molecular Formula | Metabolite | Identification Level * | Monoisotopic Mass (Da) | Mass Error (ppm) | Main Fragments (MS/MS) | VIP (Trend) ** | |
|---|---|---|---|---|---|---|---|---|---|
| Ht0.5h vs. Ct0.5h | Ht5h vs. Ct5h | ||||||||
| Intracellular fluid | |||||||||
| Significant metabolites at 5 h | |||||||||
| 1 | 0.8 | C5H7NO3 | Pyroglutamic acid | 1 | 129.0424 | 1.5 | 56.0516, 84.0469 | 0.81 (↑) | 1.19 (↑) |
| 2 | 1.8 | C11H21NO4 | Butyrylcarnitine | 1 | 231.1485 | 6.1 | 85.0283 | 0.01 (↓) | 1.51 (↑) |
| Significant metabolites at 0.5 and 5 h | |||||||||
| 3 | 0.8 | C9H17NO4 | Acetylcarnitine | 1 | 203.1188 | 14.8 | 85.0295 | 2.20 (↓) | 2.13 (↓) |
| 4 | 0.8 | C8H11NO3 | Pyridoxine | 1 | 169.0765 | 15.4 | 134.0593, 152.0705 | 1.46 (↑) | 1.39 (↑) |
| Extracellular fluid | |||||||||
| Significant metabolites at 0.5 h | |||||||||
| 5 | 5.4 | C10H11NO3 | Phenylacetylglycine | 1 | 193.0763 | 12.4 | 135.0445, 107.0516 | 1.45 (↑) | 0.59 (↑) |
| Significant metabolites at 5 h | |||||||||
| 6 | 0.9 | C12H23NO7 | N-(1-Deoxy-1-fructosyl)leucineor N-(1-deoxy-1-fructosyl)isoleucine | 2 | 293.1525 | 17.1 | 230.1378, 258.1333, 276.1420 | 0.35 (↑) | 1.12 (↑) |
| Significant metabolites at 0.5 and 5 h | |||||||||
| 7 | 3.7 | C11H15NO4 | L-4-Hydroxy-3-methoxy-a-methylphenylalanine | 2 | 225.1011 | 4.4 | 180.1008 | 1.85 (↑) | 1.46 (↑) |
| 8 | 6.4 | C15H15NO4 | Thyronine | 2 | 273.1013 | 4.4 | 228.1009 | 2.08 (↑) | 1.56 (↑) |
| PLS-DA models | R2X | R2Y | Q2 | CV-ANOVA | |
|---|---|---|---|---|---|
| Intracellular fluid | |||||
| Ht24h vs. Ct24h | 0.616 | 0.923 | 0.873 | F(39.9) | p(1.6 × 10−10) |
| Ht48h vs. Ct48h | 0.649 | 0.948 | 0.935 | F(98.8) | p(6.1 × 10−15) |
| Extracellular fluid | |||||
| Ht24h vs. Ct24h | 0.493 | 0.985 | 0.967 | F(171.3) | p(2.8 × 10−18) |
| Ht48h vs. Ct48h | 0.572 | 0.984 | 0.983 | F(776.8) | p(1.4 × 10−24) |
| # | RT (min) | Molecular Formula | Metabolite | Identification Level * | Monoisotopic Mass (Da) | Mass Error (ppm) | Main Fragments (MS/MS) | VIP (Trend) ** | |
|---|---|---|---|---|---|---|---|---|---|
| Ht24h vs. Ct24h | Ht48h vs. Ct48h | ||||||||
| Intracellular fluid | |||||||||
| Significant metabolites at 24 h | |||||||||
| 1 | 0.8 | C8H11NO3 | Pyridoxine | 1 | 169.0746 | 4.1 | 134.0597, 152.0691 | 1.16 (↓) | 0.80 (↓) |
| Significant metabolites at 48 h | |||||||||
| 2 | 0.8 | C5H7NO3 | Pyroglutamic acid | 1 | 129.0437 | 8.5 | 84.0445, 56.0491 | 0.98 (↑) | 1.25 (↑) |
| 3 | 1.3 | C9H11NO2 | Phenylalanine | 1 | 165.0781 | 5.5 | 120.0801, 103.0534 | 0.40 (↑) | 1.07 (↑) |
| Extracellular fluid | |||||||||
| Significant metabolites at 24 h | |||||||||
| 4 | 0.7 | C7H15NO3 | Carnitine | 1 | 161.1010 | 5.5 | 85.0260, 103.0327, 60.0790 | 1.16 (↓) | 0.83 (↓) |
| 5 | 0.8 | C5H11NO2 | Valine | 1 | 117.0770 | 16.2 | 72.0815, 55.0549 | 1.16 (↓) | 0.71 (↓) |
| Significant metabolites at 48 h | |||||||||
| 6 | 3.3 | C11H15NO4 | L-4-Hydroxy-3-methoxy-a-methylphenylalanine | 2 | 225.0996 | 2.2 | 180.1029 | 0.07 (↑) | 1.13 (↑) |
| 7 | 6.2 | C15H15NO4 | Thyronine | 2 | 273.0997 | 1.5 | 228.1030 | 0.51 (↑) | 1.51 (↑) |
| Significant metabolites at 24 and 48 h | |||||||||
| 8 | 0.8 | C5H7NO3 | Pyroglutamic acid | 2 | 129.0431 | 3.9 | 84.0416,56.0488 | 1.28 (↓) | 1.47 (↓) |
| 9 | 1.3 | C15H21NO7 | N-(1-deoxy-1-fructosyl)phenylalanine | 2 | 327.1295 | 7.0 | 310.1271, 292.1159, 166.0846, 178.0845 | 1.27 (↑) | 1.21 (↑) |
| 10 | 4.0 | C9H9NO3 | Hippuric acid | 1 | 179.0580 | 1.1 | 105.0327, 77.0390 | 1.27 (↑) | 1.13 (↑) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardo-Bermejo, S.; Sánchez-López, E.; Tan, L.; Benito-Martínez, S.; Jiang, Z.; Castro-Puyana, M.; Lucio-Cazaña, F.J.; Marina, M.L. Exploratory Metabolomic Analysis Based on Reversed-Phase Liquid Chromatography–Mass Spectrometry to Study an In Vitro Model of Hypoxia-Induced Metabolic Alterations in HK-2 Cells. Int. J. Mol. Sci. 2021, 22, 7399. https://doi.org/10.3390/ijms22147399
Bernardo-Bermejo S, Sánchez-López E, Tan L, Benito-Martínez S, Jiang Z, Castro-Puyana M, Lucio-Cazaña FJ, Marina ML. Exploratory Metabolomic Analysis Based on Reversed-Phase Liquid Chromatography–Mass Spectrometry to Study an In Vitro Model of Hypoxia-Induced Metabolic Alterations in HK-2 Cells. International Journal of Molecular Sciences. 2021; 22(14):7399. https://doi.org/10.3390/ijms22147399
Chicago/Turabian StyleBernardo-Bermejo, Samuel, Elena Sánchez-López, Lei Tan, Selma Benito-Martínez, Zhengjin Jiang, María Castro-Puyana, Francisco Javier Lucio-Cazaña, and María Luisa Marina. 2021. "Exploratory Metabolomic Analysis Based on Reversed-Phase Liquid Chromatography–Mass Spectrometry to Study an In Vitro Model of Hypoxia-Induced Metabolic Alterations in HK-2 Cells" International Journal of Molecular Sciences 22, no. 14: 7399. https://doi.org/10.3390/ijms22147399