Interactions of Amyloid-β with Membrane Proteins
Abstract
:1. β-Amyloid in the Development of Alzheimer’s Disease
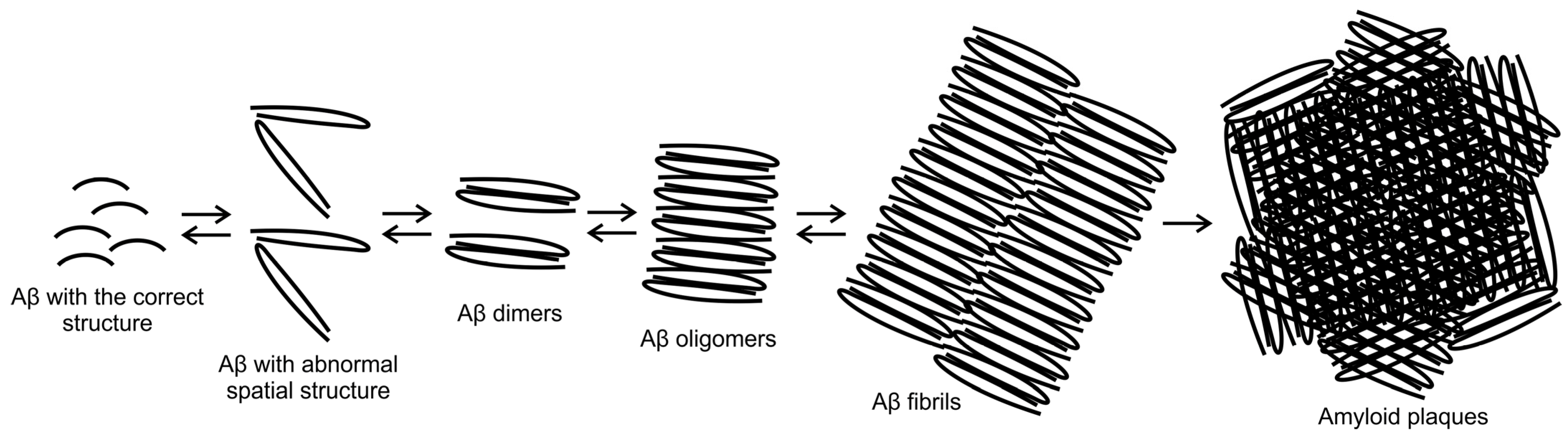
2. Structure and Cascade of β-Amyloid Transformations
3. Fragment of Aβ25–35 in the Pathomechanism of Alzheimer’s Disease
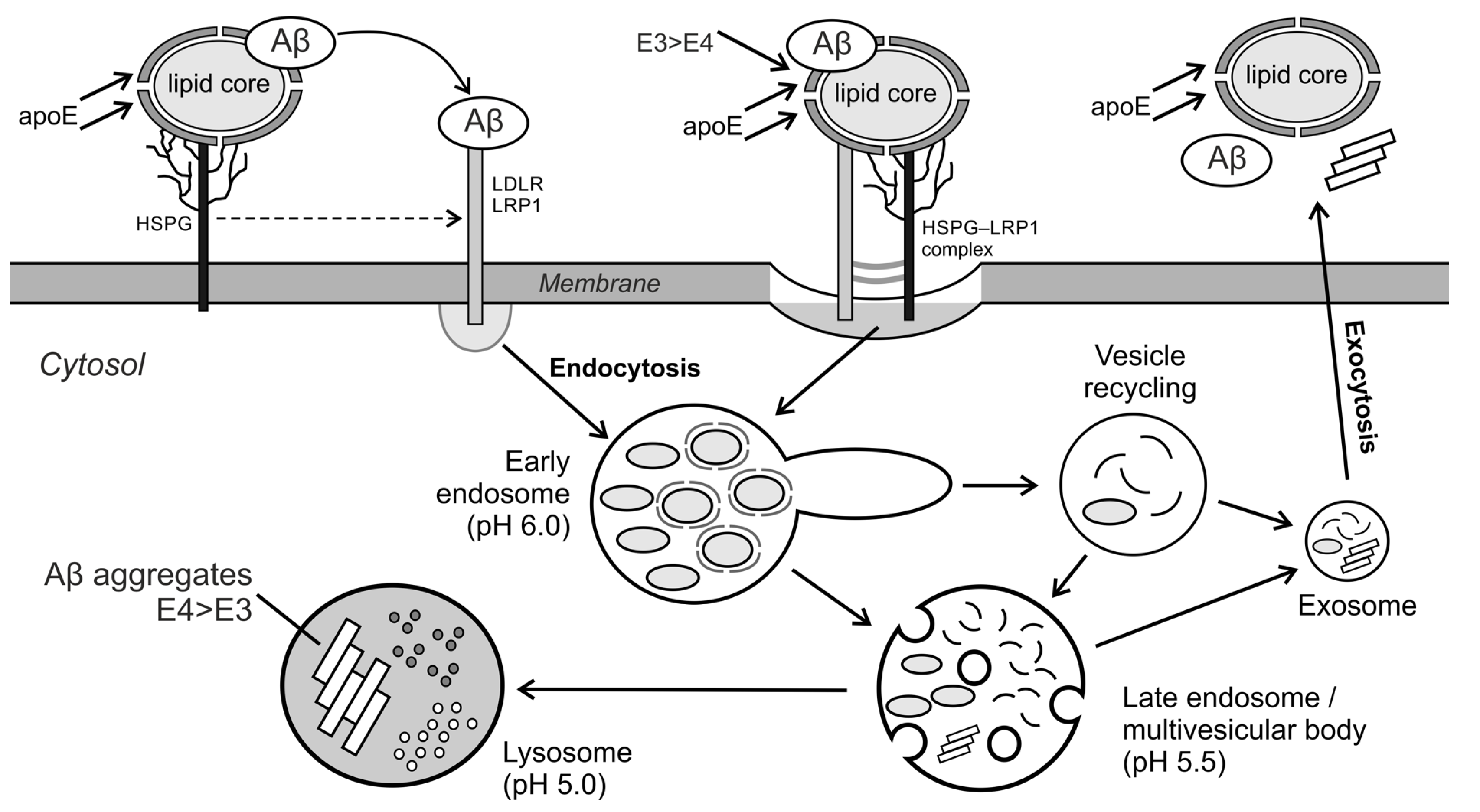
4. The Interaction of Aβ with the Cell Membrane Proteins
5. Research Methods Used to Analyze Interactions with Cell Membrane
Author Contributions
Funding
Conflicts of Interest
References
- Owen, M.C.; Kulig, W.; Poojari, C.; Rog, T.; Strodel, B. Physiologically-relevant levels of sphingomyelin, but not GM1, induces a β-sheet-rich structure in the amyloid-β(1–42) monomer. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1709–1720. [Google Scholar] [CrossRef]
- Maurya, N.; Agarwal, N.R. Neuro-pathological Implications of β-Amyloid Protein: An Update. Proc. Nat. Res. Soc. 2018, 2, 02006. [Google Scholar] [CrossRef]
- Villaflores, O.B.; Chen, Y.-J.; Chen, C.-P.; Yeh, J.-M.; Wu, T.-Y. Curcuminoids and resveratrol as anti-Alzheimer agents. Taiwan J. Obstet. Gynecol. 2012, 51, 515–525. [Google Scholar] [CrossRef] [Green Version]
- Brinkmalm, G.; Hong, W.; Wang, Z.; Liu, W.; O’Malley, T.T.; Sun, X.; Frosch, M.P.; Selkoe, D.J.; Portelius, E.; Zetterberg, H.; et al. Identification of neurotoxic cross-linked amyloid-β dimers in the Alzheimer’s brain. Brain 2019, 142, 1441–1457. [Google Scholar] [CrossRef]
- Balon, K.; Wiatrak, B. PC12 and THP-1 Cell Lines as Neuronal and Microglia Model in Neurobiological Research. Appl. Sci. 2021, 11, 3729. [Google Scholar] [CrossRef]
- Ono, K.; Condron, M.M.; Teplow, D.B. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc. Natl. Acad. Sci. USA 2009, 106, 14745–14750. [Google Scholar] [CrossRef] [Green Version]
- Penke, B.; Szucs, M.; Bogár, F. Oligomerization and conformational change turn monomeric β-amyloid and tau proteins toxic: Their role in Alzheimer’s pathogenesis. Molecules 2020, 25, 1659. [Google Scholar] [CrossRef] [Green Version]
- Dharmadana, D.; Reynolds, N.P.; Conn, C.E.; Valéry, C. Molecular interactions of amyloid nanofibrils with biological aggregation modifiers: Implications for cytotoxicity mechanisms and biomaterial design. Interface Focus 2017, 7, 20160160. [Google Scholar] [CrossRef]
- Yang, J.; Dear, A.J.; Yao, Q.Q.; Liu, Z.; Dobson, C.M.; Knowles, T.P.J.; Wu, S.; Perrett, S. Amelioration of aggregate cytotoxicity by catalytic conversion of protein oligomers into amyloid fibrils. Nanoscale 2020, 12, 18663–18672. [Google Scholar] [CrossRef]
- Niu, Z.; Zhang, Z.; Zhao, W.; Yang, J. Interactions between amyloid β peptide and lipid membranes. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1663–1669. [Google Scholar] [CrossRef]
- Gallardo, G.; Holtzman, D.M. Amyloid-β and Tau at the Crossroads of Alzheimer’s Disease. Adv. Exp. Med. Biol. 2019, 1184, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Bloom, G.S. Amyloid-β and tau: The trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurol. 2014, 71, 505–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sosa, L.J.; Cáceres, A.; Dupraz, S.; Oksdath, M.; Quiroga, S.; Lorenzo, A. The physiological role of the amyloid precursor protein as an adhesion molecule in the developing nervous system. J. Neurochem. 2017, 143, 11–29. [Google Scholar] [CrossRef] [Green Version]
- Andrew, R.J.; Fisher, K.; Heesom, K.J.; Kellett, K.A.B.; Hooper, N.M. Quantitative interaction proteomics reveals differences in the interactomes of amyloid precursor protein isoforms. J. Neurochem. 2019, 149, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Guo, T.; Zhang, D.; Zeng, Y.; Huang, T.Y.; Xu, H.; Zhao, Y. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer’s disease. Mol. Neurodegener. 2020, 15, 40. [Google Scholar] [CrossRef]
- Funamoto, S.; Tagami, S.; Okochi, M.; Morishima-Kawashima, M. Successive cleavage of β-amyloid precursor protein by γ-secretase. Semin. Cell Dev. Biol. 2020, 105, 64–74. [Google Scholar] [CrossRef]
- Nigam, S.M.; Xu, S.; Kritikou, J.S.; Marosi, K.; Brodin, L.; Mattson, M.P. Exercise and BDNF reduce Aβ production by enhancing α-secretase processing of APP. J. Neurochem. 2017, 142, 286–296. [Google Scholar] [CrossRef]
- Habekost, M.; Qvist, P.; Denham, M.; Holm, I.E.; Jørgensen, A.L. Directly Reprogrammed Neurons Express MAPT and APP Splice Variants Pertinent to Ageing and Neurodegeneration. Mol. Neurobiol. 2021, 58, 2075–2087. [Google Scholar] [CrossRef]
- Kimura, A.; Hata, S.; Suzuki, T. Alternative selection of β-site APP-cleaving enzyme 1 (BACE1) cleavage sites in amyloid β-protein precursor (APP) harboring protective and pathogenic mutations within the Aβ sequence. J. Biol. Chem. 2016, 291, 24041–24053. [Google Scholar] [CrossRef] [Green Version]
- Maltsev, A.V.; Bystryak, S.; Galzitskaya, O.V. The role of β-amyloid peptide in neurodegenerative diseases. Ageing Res. Rev. 2011, 10, 440–452. [Google Scholar] [CrossRef]
- Wolfe, M.S. Probing Mechanisms and Therapeutic Potential of γ-Secretase in Alzheimer’s Disease. Molecules 2021, 26, 388. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Mora, P.; Rogelio, L.; Colín-Barenque, L. Amyloid beta: Multiple mechanisms of toxicity and only some protective effects? Oxid. Med. Cell. Longev. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Dorandish, S.; Williams, A.; Atali, S.; Sendo, S.; Price, D.; Thompson, C.; Guthrie, J.; Heyl, D.; Evans, H.G. Regulation of amyloid-β levels by matrix metalloproteinase-2/9 (MMP2/9) in the media of lung cancer cells. Sci. Rep. 2021, 11, 9708. [Google Scholar] [CrossRef]
- Ciudad, S.; Puig, E.; Botzanowski, T.; Meigooni, M.; Arango, A.S.; Do, J.; Mayzel, M.; Bayoumi, M.; Chaignepain, S.; Maglia, G.; et al. Aβ(1–42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Quartey, M.O.; Nyarko, J.N.K.; Maley, J.M.; Barnes, J.R.; Bolanos, M.A.C.; Heistad, R.M.; Knudsen, K.J.; Pennington, P.R.; Buttigieg, J.; De Carvalho, C.E.; et al. The Aβ(1–38) peptide is a negative regulator of the Aβ(1–42) peptide implicated in Alzheimer disease progression. Sci. Rep. 2021, 11, 431. [Google Scholar] [CrossRef]
- Lattanzio, F.; Carboni, L.; Carretta, D.; Candeletti, S.; Romualdi, P. Treatment with the neurotoxic Aβ (25–35) peptide modulates the expression of neuroprotective factors Pin1, Sirtuin 1, and brain-derived neurotrophic factor in SH-SY5Y human neuroblastoma cells. Exp. Toxicol. Pathol. 2016, 68, 271–276. [Google Scholar] [CrossRef]
- Millucci, L.; Raggiaschi, R.; Franceschini, D.; Terstappen, G.; Santucci, A. Rapid aggregation and assembly in aqueous solution of A beta (25–35) peptide. J. Biosci. 2009, 34, 293–303. [Google Scholar] [CrossRef]
- Millucci, L.; Ghezzi, L.; Bernardini, G.; Santucci, A. Conformations and biological activities of amyloid beta peptide 25–35. Curr. Protein Pept. Sci. 2010, 11, 54–67. [Google Scholar] [CrossRef]
- Guo, C.-C.; Jiao, C.-H.; Gao, Z.-M. Silencing of LncRNA BDNF-AS attenuates Aβ25–35-induced neurotoxicity in PC12 cells by suppressing cell apoptosis and oxidative stress. Neurol. Res. 2018, 40, 795–804. [Google Scholar] [CrossRef]
- Gulisano, W.; Melone, M.; Ripoli, C.; Tropea, M.R.; Puma, D.D.; Giunta, S.; Cocco, S.; Marcotulli, D.; Origlia, N.; Palmeri, A.; et al. Neuromodulatory action of picomolar extracellular Aβ42 oligomers on presynaptic and postsynaptic mechanisms underlying synaptic function and memory. J. Neurosci. 2019, 39, 5986–6000. [Google Scholar] [CrossRef] [Green Version]
- Pearson, H.A.; Peers, C. Physiological roles for amyloid beta peptides. J. Physiol. 2006, 575, 5–10. [Google Scholar] [CrossRef]
- Kubo, T.; Kumagae, Y.; Miller, C.A.; Kaneko, I. β-amyloid racemized at the Ser26 residue in the brains of patients with Alzheimer disease: Implications in the pathogenesis of Alzheimer disease. J. Neuropathol. Exp. Neurol. 2003, 62, 248–259. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.P.; Perry, G.; Smith, M.A. Brain composition: Age-related changes. In Reference Module in Neuroscience and Biobehavioral Psychology; Elsevier: Amsterdam, The Netherlands, 2017; pp. 313–316. [Google Scholar]
- Tambo, K.; Yamaguchi, T.; Kobayashi, K.; Terauchi, E.; Ichi, I.; Kojo, S. Racemization of the aspartic acid residue of amyloid-β peptide by a radical reaction. Biosci. Biotechnol. Biochem. 2013, 77, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Dyakin, V.V.; Wisniewski, T.M.; Lajtha, A. Chiral interface of amyloid beta (Aβ): Relevance to protein aging, aggregation and neurodegeneration. Symmetry 2020, 12, 585. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, I.; Yamada, N.; Sakuraba, Y.; Kamenosono, M.; Tutumi, S. Suppression of mitochondrial succinate dehydrogenase, a primary target of β-amyloid, and its derivative racemized at Ser residue. J. Neurochem. 1995, 65, 2585–2593. [Google Scholar] [CrossRef]
- Chang, C.C.; Edwald, E.; Veatch, S.; Steel, D.G.; Gafni, A. Interactions of amyloid-β peptides on lipid bilayer studied by single molecule imaging and tracking. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1616–1624. [Google Scholar] [CrossRef] [PubMed]
- Meker, S.; Chin, H.; Sut, T.N.; Cho, N.J. Amyloid-β Peptide Triggers Membrane Remodeling in Supported Lipid Bilayers Depending on Their Hydrophobic Thickness. Langmuir 2018, 34, 9548–9560. [Google Scholar] [CrossRef]
- Stefani, M.; Rigacci, S. Protein folding and aggregation into amyloid: The interference by natural phenolic compounds. Int. J. Mol. Sci. 2013, 14, 12411–12457. [Google Scholar] [CrossRef] [Green Version]
- Drabik, D.; Chodaczek, G.; Kraszewski, S. Effect of amyloid-β monomers on lipid membrane mechanical parameters–potential implications for mechanically driven neurodegeneration in alzheimer’s disease. Int. J. Mol. Sci. 2021, 22, 18. [Google Scholar] [CrossRef]
- Bharadwaj, P.; Solomon, T.; Malajczuk, C.J.; Mancera, R.L.; Howard, M.; Arrigan, D.W.M.; Newsholme, P.; Martins, R.N. Role of the cell membrane interface in modulating production and uptake of Alzheimer’s beta amyloid protein. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1639–1651. [Google Scholar] [CrossRef]
- Mandal, P.K.; Pettegrew, J.W. Alzheimer’s disease: Soluble oligomeric Abeta(1–40) peptide in membrane mimic environment from solution NMR and circular dichroism studies. Neurochem. Res. 2004, 29, 2267–2272. [Google Scholar] [CrossRef]
- Fabiani, C.; Antollini, S.S. Alzheimer’s disease as a membrane disorder: Spatial cross-talk among beta-amyloid peptides, nicotinic acetylcholine receptors and lipid rafts. Front. Cell. Neurosci. 2019, 13, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokvist, M.; Lindström, F.; Watts, A.; Gröbner, G. Two types of Alzheimer’s β-amyloid (1–40) peptide membrane interactions: Aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J. Mol. Biol. 2004, 335, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Devanathan, S.; Salamon, Z.; Lindblom, G.; Grobner, G.; Tollin, G. Effects of sphingomyelin, cholesterol and zinc ions on the binding, insertion and aggregation of the amyloid Abeta1–40 peptide in solid-supported lipid bilayers. FEBS J. 2006, 273, 1389–1402. [Google Scholar] [CrossRef]
- Azouz, M.; Cullin, C.; Lecomte, S.; Lafleur, M. Membrane domain modulation of Aβ1–42 oligomer interactions with supported lipid bilayers: An atomic force microscopy investigation. Nanoscale 2019, 11, 20857–20867. [Google Scholar] [CrossRef]
- Lai, A.Y.; McLaurin, J. Mechanisms of amyloid-beta peptide uptake by neurons: The role of lipid rafts and lipid raft-associated proteins. Int. J. Alzheimer’s Dis. 2010, 2011. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, K. Pathological significance of ganglioside clusters in Alzheimer’s disease. J. Neurochem. 2011, 116, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Ahyayauch, H.; Masserini, M.; Goñi, F.M.; Alonso, A. The interaction of Aβ42 peptide in monomer, oligomer or fibril forms with sphingomyelin/cholesterol/ganglioside bilayers. Int. J. Biol. Macromol. 2021, 168, 611–619. [Google Scholar] [CrossRef]
- Cebecauer, M.; Hof, M.; Amaro, M. Impact of GM1 on Membrane-Mediated Aggregation/Oligomerization of β-Amyloid: Unifying View. Biophys. J. 2017, 113, 1194–1199. [Google Scholar] [CrossRef] [Green Version]
- Amaro, M.; Šachl, R.; Aydogan, G.; Mikhalyov, I.I.; Vácha, R.; Hof, M. GM1 ganglioside inhibits β-amyloid oligomerization induced by sphingomyelin. Angew. Chem. Int. Ed. 2016, 55, 9411–9415. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Wang, Q.; Min, L.; Sui, R.; Li, J.; Liu, X. Monosialoanglioside improves memory deficits and relieves oxidative stress in the hippocampus of rat model of Alzheimer’s disease. Neurol. Sci. 2013, 34, 1447–1451. [Google Scholar] [CrossRef] [PubMed]
- Kreutz, F.; Scherer, E.B.; Ferreira, A.G.; dos Santos Petry, F.; Pereira, C.L.; Santana, F.; de Souza Wyse, A.T.; Salbego, C.G.; Trindade, V.M.T. Alterations on Na+,K+-ATPase and acetylcholinesterase activities induced by amyloid-β peptide in rat brain and GM1 ganglioside neuroprotective action. Neurochem. Res. 2013, 38, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Mulaj, M.; Foley, J.; Muschol, M. Amyloid oligomers and protofibrils, but not filaments, self-replicate from native lysozyme. J. Am. Chem. Soc. 2014, 136, 8947–8956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.F.; Xu, T.H.; Yan, Y.; Zhou, Y.R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef] [PubMed]
- Lau, T.-L.; Gehman, J.D.; Wade, J.D.; Perez, K.; Masters, C.L.; Barnham, K.J.; Separovic, F. Membrane interactions and the effect of metal ions of the amyloidogenic fragment Aβ(25–35) in comparison to Aβ(1–42). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2400–2408. [Google Scholar] [CrossRef] [Green Version]
- Ureshino, R.P.; Erustes, A.G.; Bassani, T.B.; Wachilewski, P.; Guarache, G.C.; Nascimento, A.C.; Costa, A.J.; Smaili, S.S.; da Silva Pereira, G.J. The interplay between ca2+ signaling pathways and neurodegeneration. Int. J. Mol. Sci. 2019, 20, 6004. [Google Scholar] [CrossRef] [Green Version]
- Jaudon, F.; Thalhammer, A.; Cingolani, L.A. Integrin adhesion in brain assembly: From molecular structure to neuropsychiatric disorders. Eur. J. Neurosci. 2020. [Google Scholar] [CrossRef]
- Picone, P.; Nuzzo, D.; Giacomazza, D.; Di Carlo, M. β-Amyloid Peptide: The Cell Compartment Multi-faceted Interaction in Alzheimer’s Disease. Neurotox. Res. 2020, 37, 250–263. [Google Scholar] [CrossRef]
- Forester, B.P.; Berlow, Y.A.; Harper, D.G.; Jensen, J.E.; Lange, N.; Froimowitz, M.P.; Ravichandran, C.; Iosifescu, D.V.; Lukas, S.E.; Renshaw, P.F.; et al. Age-related changes in brain energetics and phospholipid metabolism. NMR Biomed. 2009, 23, 242–250. [Google Scholar] [CrossRef]
- Kanekiyo, T.; Xu, H.; Bu, G. ApoE and Aβ in Alzheimer’s disease: Accidental encounters or partners? Neuron 2014, 81, 740–754. [Google Scholar] [CrossRef] [Green Version]
- Marshall, K.E.; Vadukul, D.M.; Staras, K.; Serpell, L.C. Misfolded amyloid-β-42 impairs the endosomal–lysosomal pathway. Cell. Mol. Life Sci. 2020, 77, 5031–5043. [Google Scholar] [CrossRef] [Green Version]
- Han, K.; Kim, S.H.; Choi, M. Computational modeling of the effects of autophagy on amyloid-β peptide levels. Theor. Biol. Med. Model. 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, H.B. Targeting Fyn kinase in Alzheimer’s disease. Biol. Psychiatry 2018, 83, 369–376. [Google Scholar] [CrossRef]
- Larson, M.; Sherman, M.A.; Amar, F.; Nuvolone, M.; Schneider, J.A.; Bennett, D.A.; Aguzzi, A.; Lesné, S.E.; Lesne, S.E. The complex PrP(c)-Fyn couples human oligomeric Aβ with pathological tau changes in Alzheimer’s disease. J. Neurosci. 2012, 32, 16857–16871. [Google Scholar] [CrossRef] [Green Version]
- Ittner, L.M.; Ke, Y.D.; Delerue, F.; Bi, M.; Gladbach, A.; van Eersel, J.; Wölfing, H.; Chieng, B.C.; Christie, M.J.; Napier, I.A.; et al. Dendritic function of tau mediates amyloid-β toxicity in alzheimer’s disease mouse models. Cell 2010, 142, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haass, C.; Mandelkow, E. Fyn-Tau-Amyloid: A toxic triad. Cell 2010, 142, 356–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Yang, G.; Shi, Y. Macromolecular complex in recognition and proteolysis of amyloid precursor protein in Alzheimer’s disease. Curr. Opin. Struct. Biol. 2020, 61, 1–8. [Google Scholar] [CrossRef]
- Wärmländer, S.K.T.S.; Österlund, N.; Wallin, C.; Wu, J.; Luo, J.; Tiiman, A.; Jarvet, J.; Gräslund, A. Metal binding to the amyloid-β peptides in the presence of biomembranes: Potential mechanisms of cell toxicity. J. Biol. Inorg. Chem. 2019, 24, 1189–1196. [Google Scholar] [CrossRef] [Green Version]
- Bagheri, Y.; Ali, A.A.; You, M. Current Methods for Detecting Cell Membrane Transient Interactions. Front. Chem. 2020, 8, 1074. [Google Scholar] [CrossRef]
- Ask, M.; Reza, M. Computational Models in Neuroscience: How real are they? A Critical Review of Status and Suggestions. Austin Neurol Neurosci. 2016, 1, 1008. [Google Scholar]
- Yeh, V.; Goode, A.; Bonev, B.B. Membrane protein structure determination and characterisation by solution and solid-state nmr. Biology 2020, 9, 396. [Google Scholar] [CrossRef]
- Bender, J.; Schmidt, C. Mass spectrometry of membrane protein complexes. Biol. Chem. 2019, 400, 813–829. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Moro, M.; Di Silvio, D.; Moya, S.E. Fluorescence correlation spectroscopy as a tool for the study of the intracellular dynamics and biological fate of protein corona. Biophys. Chem. 2019, 253, 106218. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.M.; Gaus, K. Imaging lipid domains in cell membranes: The advent of super-resolution fluorescence microscopy. Front. Plant Sci. 2013, 4, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patrício, D.; Fardilha, M. The mammalian two-hybrid system as a powerful tool for high-throughput drug screening. Drug Discov. Today 2020, 25, 764–771. [Google Scholar] [CrossRef]
- Stasi, M.; De Luca, M.; Bucci, C. Two-hybrid-based systems: Powerful tools for investigation of membrane traffic machineries. J. Biotechnol. 2015, 202, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, Y.; Chedid, S.; Shafiei, F.; Zhao, B.; You, M. A quantitative assessment of the dynamic modification of lipid-DNA probes on live cell membranes. Chem. Sci. 2019, 10, 11030–11040. [Google Scholar] [CrossRef] [PubMed]
- Hedegaard, S.F.; Derbas, M.S.; Lind, T.K.; Kasimova, M.R.; Christensen, M.V.; Michaelsen, M.H.; Campbell, R.A.; Jorgensen, L.; Franzyk, H.; Cárdenas, M.; et al. Fluorophore labeling of a cell-penetrating peptide significantly alters the mode and degree of biomembrane interaction. Sci. Rep. 2018, 8, 6327. [Google Scholar] [CrossRef] [PubMed]
- Feuillie, C.; Lambert, E.; Ewald, M.; Azouz, M.; Henry, S.; Marsaudon, S.; Cullin, C.; Lecomte, S.; Molinari, M. High Speed AFM and NanoInfrared Spectroscopy Investigation of Aβ1–42 Peptide Variants and Their Interaction With POPC/SM/Chol/GM1 Model Membranes. Front. Mol. Biosci. 2020, 7, 238. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiatrak, B.; Piasny, J.; Kuźniarski, A.; Gąsiorowski, K. Interactions of Amyloid-β with Membrane Proteins. Int. J. Mol. Sci. 2021, 22, 6075. https://doi.org/10.3390/ijms22116075
Wiatrak B, Piasny J, Kuźniarski A, Gąsiorowski K. Interactions of Amyloid-β with Membrane Proteins. International Journal of Molecular Sciences. 2021; 22(11):6075. https://doi.org/10.3390/ijms22116075
Chicago/Turabian StyleWiatrak, Benita, Janusz Piasny, Amadeusz Kuźniarski, and Kazimierz Gąsiorowski. 2021. "Interactions of Amyloid-β with Membrane Proteins" International Journal of Molecular Sciences 22, no. 11: 6075. https://doi.org/10.3390/ijms22116075





