Mining the Biomarker Potential of the Urine Peptidome: From Amino Acids Properties to Proteases
Abstract
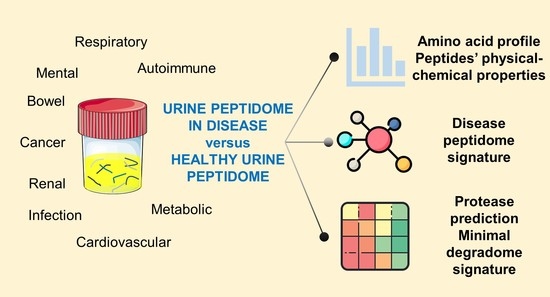
:1. Introduction
The Potential of Urine Peptidomics
2. Brief Methods
2.1. Literature Search and Data Mining
2.2. Statistical and Bioinformatics Analyses
3. Results and Discussion
3.1. The Urinary Peptidome as a Road to Defining Molecular Disease Signatures
3.2. Can We Distinguish Disease Classes by Peptides’ Physical-Chemical Properties?
3.3. Can Proteases Help Distinguish Diseases?
4. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Caron, E.; Aebersold, R.; Banaei-Esfahani, A.; Chong, C.; Bassani-Sternberg, M. A Case for a human immuno-peptidome project consortium. Immunity 2017, 47, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Gelman, J.S.; Fricker, L.D. Hemopressin and other bioactive peptides from cytosolic proteins: Are these non-classical neuropeptides? AAPS J. 2010, 12, 279–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, P.Y.; Low, T.Y.; Jamal, R. Probing the endogenous peptidomes of cancer for biomarkers: A new endeavor. In Advances in Clinical Chemistry; Elsevier: Cambridge, MA, USA, 2019; pp. 67–89. ISBN 0065-2423. [Google Scholar]
- Sarkizova, S.; Klaeger, S.; Le, P.M.; Li, L.W.; Oliveira, G.; Keshishian, H.; Hartigan, C.R.; Zhang, W.; Braun, D.A.; Ligon, K.L.; et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 2020, 38, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Lygirou, V.; Latosinska, A.; Makridakis, M.; Mullen, W.; Delles, C.; Schanstra, J.P.; Zoidakis, J.; Pieske, B.; Mischak, H.; Vlahou, A. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J. Transl. Med. 2018, 16, 104. [Google Scholar] [CrossRef]
- Puente, X.S.; Sánchez, L.M.; Overall, C.M.; López-Otín, C. Human and mouse proteases: A comparative genomic approach. Nat. Rev. Genet. 2003, 4, 544. [Google Scholar] [CrossRef]
- Lee, P.Y.; Osman, J.; Low, T.Y.; Jamal, R. Plasma/Serum proteomics: Depletion strategies for reducing high-abundance proteins for biomarker discovery. Bioanalysis 2019, 11, 1799–1812. [Google Scholar] [CrossRef]
- Golubnitschaja, O.; Baban, B.; Boniolo, G.; Wang, W.; Bubnov, R.; Kapalla, M.; Krapfenbauer, K.; Mozaffari, M.S.; Costigliola, V. Medicine in the early twenty-first century: Paradigm and anticipation—EPMA position paper 2016. EPMA J. 2016, 7, 23. [Google Scholar] [CrossRef] [Green Version]
- Latosinska, A.; Siwy, J.; Mischak, H.; Frantzi, M. Peptidomics and proteomics based on CE-MS as a robust tool in clinical application: The past, the present, and the future. Electrophoresis 2019, 40, 2294–2308. [Google Scholar] [CrossRef]
- Di Meo, A.; Pasic, M.D.; Yousef, G.M. Proteomics and peptidomics: Moving toward precision medicine in urological malignancies. Oncotarget 2016, 7, 52460–52474. [Google Scholar] [CrossRef] [Green Version]
- Greening, D.W.; Kapp, E.A.; Simpson, R.J. The peptidome comes of age: Mass spectrometry-based characterization of the circulating cancer peptidome. Pept. Cancer Deriv. Enzym. Prod. 2017, 42, 27–64. [Google Scholar] [CrossRef]
- Schrader, M. Origins, technological development, and applications of peptidomics. In Peptidomics; Schrader, M., Fricker, L., Eds.; Springer: New York, NY, USA, 2018; pp. 3–39. ISBN 978-1-4939-7537-2. [Google Scholar]
- Cai, T.; Yang, F. Strategies for characterization of low-abundant intact or truncated low-molecular-weight proteins from human plasma. Enzymes 2017, 42, 105–123. [Google Scholar] [CrossRef]
- Klein, J.; Bascands, J.-L.; Mischak, H.; Schanstra, J.P. The role of urinary peptidomics in kidney disease research. Kidney Int. 2016, 89, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Štěpánová, S.; Kašička, V. Analysis of proteins and peptides by electromigration methods in microchips. J. Sep. Sci. 2016, 40, 228–250. [Google Scholar] [CrossRef]
- Pontillo, C.; Mischak, H. Urinary peptide-based classifier CKD273: Towards clinical application in chronic kidney disease. Clin. Kidney J. 2017, 10, 192–201. [Google Scholar] [CrossRef]
- Øverbye, A.; Skotland, T.; Koehler, C.J.; Thiede, B.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015, 6, 30357–30376. [Google Scholar] [CrossRef] [Green Version]
- Markoska, A.; Valaiyapathi, R.; Thorn, C.; Dornhorst, A. Urinary C peptide creatinine ratio in pregnant women with normal glucose tolerance and type 1 diabetes: Evidence for insulin secretion. BMJ Open Diabetes Res. Care 2017, 5, e000313. [Google Scholar] [CrossRef] [Green Version]
- Schanstra, J.P.; Zürbig, P.; Alkhalaf, A.; Argiles, A.; Bakker, S.J.L.; Beige, J.; Bilo, H.J.G.; Chatzikyrkou, C.; Dakna, M.; Dawson, J.; et al. Diagnosis and prediction of CKD progression by assessment of urinary peptides. J. Am. Soc. Nephrol. 2015, 26, 1999–2010. [Google Scholar] [CrossRef]
- Pontillo, C.; Mischak, H. Urinary biomarkers to predict CKD: Is the future in multi-marker panels? Nephrol. Dial. Transplant. 2016, 31, 1373–1375. [Google Scholar] [CrossRef] [Green Version]
- Tofte, N.; Lindhardt, M.; Adamova, K.; Bakker, S.J.L.; Beige, J.; Beulens, J.W.J.; Birkenfeld, A.L.; Currie, G.; Delles, C.; Dimos, I.; et al. Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): A prospective observational study and embedded randomised placebo-controlled trial. Lancet Diabetes Endocrinol. 2020, 8, 301–312. [Google Scholar] [CrossRef]
- Markoska, K.; Pejchinovski, M.; Pontillo, C.; Zürbig, P.; Dakna, M.; Masin-Spasovska, J.; Stojceva-Taneva, O.; Mischak, H.; Spasovski, G. MO024 urinary peptide biomarkers associated with improvement in eGFR in CKD patients. Nephrol. Dial. Transplant. 2016, 31, i38. [Google Scholar] [CrossRef] [Green Version]
- Van, J.A.D.; Scholey, J.W.; Konvalinka, A. Insights into diabetic kidney disease using urinary proteomics and bioinformatics. J. Am. Soc. Nephrol. 2017, 28, 1050–1061. [Google Scholar] [CrossRef]
- Krochmal, M.; Kontostathi, G.; Magalhães, P.; Makridakis, M.; Klein, J.; Husi, H.; Leierer, J.; Mayer, G.; Bascands, J.-L.; Denis, C.; et al. Urinary peptidomics analysis reveals proteases involved in diabetic nephropathy. Sci. Rep. 2017, 7, 15160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustamante, C.J.; Kaiser, C.M.; Maillard, R.A.; Goldman, D.H.; Wilson, C.A.M. Mechanisms of cellular proteostasis: Insights from single-molecule approaches. Annu. Rev. Biophys. 2014, 43, 119–140. [Google Scholar] [CrossRef] [Green Version]
- Andersen, H.; Friis, U.G.; Hansen, P.B.L.; Svenningsen, P.; Henriksen, J.E.; Jensen, B.L. Diabetic nephropathy is associated with increased urine excretion of proteases plasmin, prostasin and urokinase and activation of amiloride-sensitive current in collecting duct cells. Nephrol. Dial. Transplant. 2015, 30, 781–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhusudhan, T.; Kerlin, B.A.; Isermann, B. The emerging role of coagulation proteases in kidney disease. Nat. Rev. Nephrol. 2016, 12, 94–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Boer, I.H. A New chapter for diabetic kidney disease. N. Engl. J. Med. 2017, 377, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Koya, D.; Kanasaki, K. Dipeptidyl peptidase-4 and kidney fibrosis in diabetes. Fibrogenes. Tissue Repair 2016, 9, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padoan, A.; Basso, D.; Zambon, C.-F.; Prayer-Galetti, T.; Arrigoni, G.; Bozzato, D.; Moz, S.; Zattoni, F.; Bellocco, R.; Plebani, M. MALDI-TOF peptidomic analysis of serum and post-prostatic massage urine specimens to identify prostate cancer biomarkers. Clin. Proteom. 2018, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Chinello, C.; Cazzaniga, M.; De Sio, G.; Smith, A.J.; Gianazza, E.; Grasso, A.; Rocco, F.; Signorini, S.; Grasso, M.; Bosari, S.; et al. Urinary signatures of renal cell carcinoma investigated by peptidomic approaches. PLoS ONE 2014, 9, e106684. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, P.; Pontillo, C.; Pejchinovski, M.; Siwy, J.; Krochmal, M.; Makridakis, M.; Carrick, E.; Klein, J.; Mullen, W.; Jankowski, J.; et al. Comparison of urine and plasma peptidome indicates selectivity in renal peptide handling. Proteom. Clin. Appl. 2018, 12, 1700163. [Google Scholar] [CrossRef]
- Sirolli, V.; Pieroni, L.; Di Liberato, L.; Urbani, A.; Bonomini, M. Urinary peptidomic biomarkers in kidney diseases. Int. J. Mol. Sci. 2019, 21, 96. [Google Scholar] [CrossRef] [Green Version]
- Pejchinovski, M.; Siwy, J.; Mullen, W.; Mischak, H.; Petri, M.A.; Burkly, L.C.; Wei, R. Urine peptidomic biomarkers for diagnosis of patients with systematic lupus erythematosus. Lupus 2017, 27, 6–16. [Google Scholar] [CrossRef] [Green Version]
- Nkuipou-Kenfack, E.; Bhat, A.; Klein, J.; Jankowski, V.; Mullen, W.; Vlahou, A.; Dakna, M.; Koeck, T.; Schanstra, J.P.; Zürbig, P.; et al. Identification of ageing-associated naturally occurring peptides in human urine. Oncotarget 2015, 6, 34106–34117. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Ravassa, S.; Nkuipou-Kenfack, E.; Yang, W.-Y.; Kerr, S.M.; Koeck, T.; Campbell, A.; Kuznetsova, T.; Mischak, H.; Padmanabhan, S.; et al. Novel urinary peptidomic classifier predicts incident heart failure. J. Am. Heart Assoc. 2017, 6, e005432. [Google Scholar] [CrossRef]
- Siwy, J.; Mullen, W.; Golovko, I.; Franke, J.; Zürbig, P. Human urinary peptide database for multiple disease biomarker discovery. Proteom. Clin. Appl. 2011, 5, 367–374. [Google Scholar] [CrossRef]
- Di Meo, A.; Bartruch, I.; Yousef, A.G.; Pasic, M.D.; Diamandis, E.P.; Yousef, G.M. An integrated proteomic and peptidomic assessment of the normal human urinome. Clin. Chem. Lab. Med. 2017, 55, 237. [Google Scholar] [CrossRef]
- Osorio, D.; Rondon-Villarreal, P.; Torres, R. Peptides: A package for data mining of antimicrobial peptides. R J. 2015, 7, 4–14. [Google Scholar] [CrossRef]
- Klein, J.; Eales, J.; Zürbig, P.; Vlahou, A.; Mischak, H.; Stevens, R. Proteasix: A tool for automated and large-scale prediction of proteases involved in naturally occurring peptide generation. Proteomics 2013, 13, 1077–1082. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Frantzi, M.; Metzger, J.; Banks, R.E.; Husi, H.; Klein, J.; Dakna, M.; Mullen, W.; Cartledge, J.J.; Schanstra, J.P.; Brand, K.; et al. Discovery and validation of urinary biomarkers for detection of renal cell carcinoma. J. Proteom. 2014, 98, 44–58. [Google Scholar] [CrossRef]
- Van, J.A.D.; Clotet-Freixas, S.; Zhou, J.; Batruch, I.; Sun, C.; Glogauer, M.; Rampoldi, L.; Elia, Y.; Mahmud, F.H.; Sochett, E.; et al. Peptidomic analysis of urine from youths with early type 1 diabetes reveals novel bioactivity of uromodulin peptides in vitro. Mol. Cell. Proteom. 2020, 19, 501–517. [Google Scholar] [CrossRef]
- Martin-Lorenzo, M.; Zubiri, I.; Maroto, A.S.; Gonzalez-Calero, L.; Posada-Ayala, M.; de la Cuesta, F.; Mourino-Alvarez, L.; Lopez-Almodovar, L.F.; Calvo-Bonacho, E.; Ruilope, L.M.; et al. KLK1 and ZG16B proteins and arginine-proline metabolism identified as novel targets to monitor atherosclerosis, acute coronary syndrome and recovery. Metabolomics 2015, 11, 1056–1067. [Google Scholar] [CrossRef] [Green Version]
- Gobin, E.; Bagwell, K.; Wagner, J.; Mysona, D.; Sandirasegarane, S.; Smith, N.; Bai, S.; Sharma, A.; Schleifer, R.; She, J.-X. A pan-cancer perspective of matrix metalloproteases (MMP) gene expression profile and their diagnostic/prognostic potential. BMC Cancer 2019, 19, 581. [Google Scholar] [CrossRef] [Green Version]
- Oe, Y.; Hayashi, S.; Fushima, T.; Sato, E.; Kisu, K.; Sato, H.; Ito, S.; Takahashi, N. Coagulation factor Xa and protease-activated receptor 2 as novel therapeutic targets for diabetic nephropathy. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1525–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohl, D.L.; Brennan, D.C. BK virus nephropathy and kidney transplantation. Clin. J. Am. Soc. Nephrol. 2007, 2, S36–S46. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Kim, J.; Stanley, M.; He, Z.; Faubel, S.; Schrier, R.W.; Edelstein, C.L. Pathways of caspase-mediated apoptosis in autosomal-dominant polycystic kidney disease (ADPKD). Kidney Int. 2005, 67, 909–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, Z.; Al-Lamki, R.; Wang, J.; Zuo, N.; Bradley, J.R.; Wang, L. The role of tumor necrosis factor-α converting enzyme in renal transplant rejection. Am. J. Nephrol. 2010, 32, 362–368. [Google Scholar] [CrossRef]
- McKerrow, J.H.; Caffrey, C.; Kelly, B.; Loke, P.; Sajid, M. Proteases in parasitic diseases. Annu. Rev. Pathol. Mech. Dis. 2006, 1, 497–536. [Google Scholar] [CrossRef]
- Gerner, C.; Costigliola, V.; Golubnitschaja, O. Multiomic patterns in body fluids: Technological challenge with a great potential to implement the advanced paradigm of 3P medicine. Mass Spectrom. Rev. 2020, 39, 442–451. [Google Scholar] [CrossRef]






| Condition 1 | Class | Minimal Degradome Signature |
|---|---|---|
| Type 2 diabetic nephropathy | Renal | F10 2 |
| BK virus nephritis | Infection | GZMK or TPSAB1 2 |
| End-stage renal disease in the setting of autosomal dominant polycystic kidney disease | Renal | CASP2 or CASP8 |
| Necrotizing enterocolitis | Bowel | GZMB |
| Acute rejection of kidney transplant | Autoimmune | ADAM17 |
| Schistosoma haematobium infection | Infection | PGA3 |
| Major depressive disorder | Mental | PCSK2, HTRA2 or CELA1 |
| Acute Kawasaki disease | Cardiovascular | CAPN1 + MMP7 |
| Bladder cancer | Cancer | CTSE + MCPT3 |
| Lupus nephritis | Renal | PITRM1 + PGC |
| Renal cell cancer | Cancer | KLK3 + CTSK |
| Preeclampsia | Cardiovascular | CTSK + BMP1 |
| Diabetes mellitus | Metabolic | MMP17 + BMP1 |
| Autosomal dominant polycystic kidney disease | Renal | KLK6 + MMP9 |
| Left ventricular diastolic dysfunction and hypertension | Cardiovascular | BMP1 + TMPRSS11D |
| Prostate cancer | Cancer | CTSE + MMP2 |
| Helicobacter pylori infection | Infection | ADAMTS4 + KLK4 |
| Diabetic nephropathy versus chronic renal disease | Renal | GZMA + MMP10 |
| Acute kidney injury | Renal | (CASP3 or CASP6) + (ADAMTS4 or MMP2) |
| Anti-neutrophil cytoplasmic antibody-associated vasculitis | Autoimmune | MMP17 + CTSD + PGC |
| Type 2 diabetes mellitus | Metabolic | CAPN1 + CAPN2 + ELANE |
| Type 1 diabetes mellitus | Metabolic | CTSK + ADAM10 + CASP1 |
| Systemic juvenile idiopathic arthritis | Autoimmune | ADAM10 + CASP1 + F2 |
| Left ventricular diastolic dysfunction | Cardiovascular | MMP9 + TMPRSS11D + THOP1 |
| Chronic kidney disease | Renal | ADAMTS4 + MMP9 + MMP25 |
| Type 1 diabetes mellitus versus Type 2 diabetes mellitus | Metabolic | KLK14 + KLK2 + MMP3 |
| Chronic obstructive pulmonary disease with alpha-1-antitrypsin deficiency | Respiratory | TMPRS11D + KLK6 + NLN |
| Chronic allograft nephropathy or dysfunction | Renal | GZMA + PCSK5 + (F2 or TMPRS11D) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trindade, F.; Barros, A.S.; Silva, J.; Vlahou, A.; Falcão-Pires, I.; Guedes, S.; Vitorino, C.; Ferreira, R.; Leite-Moreira, A.; Amado, F.; et al. Mining the Biomarker Potential of the Urine Peptidome: From Amino Acids Properties to Proteases. Int. J. Mol. Sci. 2021, 22, 5940. https://doi.org/10.3390/ijms22115940
Trindade F, Barros AS, Silva J, Vlahou A, Falcão-Pires I, Guedes S, Vitorino C, Ferreira R, Leite-Moreira A, Amado F, et al. Mining the Biomarker Potential of the Urine Peptidome: From Amino Acids Properties to Proteases. International Journal of Molecular Sciences. 2021; 22(11):5940. https://doi.org/10.3390/ijms22115940
Chicago/Turabian StyleTrindade, Fábio, António S. Barros, Jéssica Silva, Antonia Vlahou, Inês Falcão-Pires, Sofia Guedes, Carla Vitorino, Rita Ferreira, Adelino Leite-Moreira, Francisco Amado, and et al. 2021. "Mining the Biomarker Potential of the Urine Peptidome: From Amino Acids Properties to Proteases" International Journal of Molecular Sciences 22, no. 11: 5940. https://doi.org/10.3390/ijms22115940