Kaempferol and Its Glycoside, Kaempferol 7-O-rhamnoside, Inhibit PD-1/PD-L1 Interaction In Vitro
Abstract
:1. Introduction
2. Results
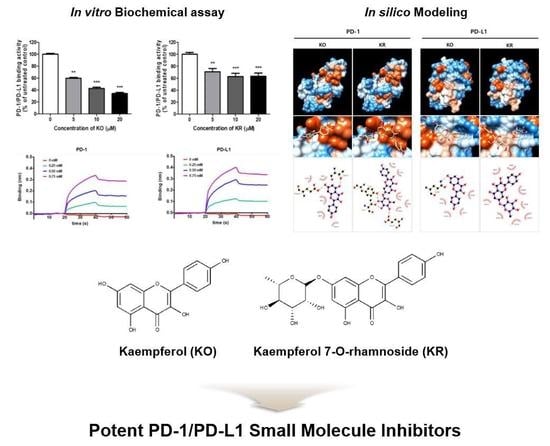
2.1. Effects of KO and Its Glycosides on PD-1/PD-L1 Protein Interaction
2.2. Effects of KO and Its Glycosides on PD-1/PD-L1 Interaction in a Cell Model System
2.3. The Binding Interaction and Affinity of KR with PD-1 and PD-L1
2.4. KO and Its Glycosides Inhibit PD-1/PD-L1 Protein Interaction In Silico
3. Discussion
4. Materials and Methods
4.1. Chemicals and Antibodies
4.2. Preparation of GHE
4.3. Competitive Enzyme-Linked Immunosorbent Assay (ELISA)
4.4. Cell Culture
4.5. PD-1/PD-L1 Blockade Assay
4.6. Kinetic Analysis by Biolayer Interferometry (BLI Analysis)
4.7. In Silico Docking Simulation and Interaction Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed death-ligand 1 |
| TCR | T-cell receptor |
| IFN-γ | Interferon-gamma |
| NFAT | Nuclear factor of activated T cells |
| IL-2 | Interleukin 2 |
| ICB | Immune checkpoint blockade |
| SPR | Surface Plasmon Resonance |
| BLI | Biolayer Interferometry |
| KO | Kaempferol |
| KR | Kaempferol-7-O-rhamnose |
| KI | Kaempferitrin, kaempferol-3,7-dirhamnoside |
References
- Fife, B.T.; Pauken, K.E. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann. N. Y. Acad. Sci. 2011, 1217, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Guzik, K.; Tomala, M.; Muszak, D.; Konieczny, M.; Hec, A.; Blaszkiewicz, U.; Pustula, M.; Butera, R.; Domling, A.; Holak, T.A. Development of the Inhibitors that Target the PD-1/PD-L1 Interaction-A Brief Look at Progress on Small Molecules, Peptides and Macrocycles. Molecules 2019, 24, 2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oestreich, K.J.; Yoon, H.; Ahmed, R.; Boss, J.M. NFATc1 regulates PD-1 expression upon T cell activation. J. Immunol. 2008, 181, 4832–4839. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Hamanishi, J.; Chamoto, K.; Honjo, T. Cancer immunotherapies targeting the PD-1 signaling pathway. J. Biomed. Sci. 2017, 24, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, R.; Sugiura, D.; Shimizu, K.; Maruhashi, T.; Watada, M.; Okazaki, I.M.; Okazaki, T. PD-1 Primarily Targets TCR Signal in the Inhibition of Functional T Cell Activation. Front. Immunol. 2019, 10, 630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serfling, E.; Klein-Hessling, S.; Palmetshofer, A.; Bopp, T.; Stassen, M.; Schmitt, E. NFAT transcription factors in control of peripheral T cell tolerance. Eur. J. Immunol. 2006, 36, 2837–2843. [Google Scholar] [CrossRef]
- Chen, L.; Flies, D.B. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013, 13, 227–242. [Google Scholar] [CrossRef]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget 2017, 8, 72167–72181. [Google Scholar] [CrossRef] [Green Version]
- Alsaab, H.O.; Sau, S.; Alzhrani, R.; Tatiparti, K.; Bhise, K.; Kashaw, S.K.; Iyer, A.K. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front. Pharmacol. 2017, 8, 561. [Google Scholar] [CrossRef]
- Borchers, A.T.; Hackman, R.M.; Keen, C.L.; Stern, J.S.; Gershwin, M.E. Complementary medicine: A review of immunomodulatory effects of Chinese herbal medicines. Am. J. Clin. Nutr. 1997, 66, 1303–1312. [Google Scholar] [CrossRef] [Green Version]
- Kwon, D.H.; Cheon, J.M.; Choi, E.O.; Jeong, J.W.; Lee, K.W.; Kim, K.Y.; Kim, S.G.; Kim, S.; Hong, S.H.; Park, C.; et al. The Immunomodulatory Activity of Mori folium, the Leaf of Morus alba L. in RAW 264.7 Macrophages In Vitro. J. Cancer Prev. 2016, 21, 144–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.D.; Deng, Y.R.; Tian, Z.G.; Lian, Z.X. Traditional Chinese Medicine and Immune Regulation. Clin. Rev. Allerg. Immu. 2013, 44, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.K.H.; Vanitha, J. Immunomodulatory and antimicrobial effects of some traditional Chinese medicinal herbs: A review. Curr. Med. Chem. 2004, 11, 1423–1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceci, C.; Tentori, L.; Atzori, M.G.; Lacal, P.M.; Bonanno, E.; Scimeca, M.; Cicconi, R.; Mattei, M.; de Martino, M.G.; Vespasiani, G.; et al. Ellagic Acid Inhibits Bladder Cancer Invasiveness and In Vivo Tumor Growth. Nutrients 2016, 8, 744. [Google Scholar] [CrossRef] [PubMed]
- Coombs, M.R.; Harrison, M.E.; Hoskin, D.W. Apigenin inhibits the inducible expression of programmed death ligand 1 by human and mouse mammary carcinoma cells. Cancer Lett. 2016, 380, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Rawangkan, A.; Wongsirisin, P.; Namiki, K.; Iida, K.; Kobayashi, Y.; Shimizu, Y.; Fujiki, H.; Suganuma, M. Green Tea Catechin Is an Alternative Immune Checkpoint Inhibitor that Inhibits PD-L1 Expression and Lung Tumor Growth. Molecules 2018, 23, 2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Mejia, E.G.; Mazewski, C.; Luna-Vital, D.; Berhow, M. Black Lentil Aqueous Extract Attenuates Colitis-Associated Colon Carcinogenesis in Mice (OR04-06-19). Curr. Dev. Nutr. 2019, 3, OR04-06-19. [Google Scholar] [CrossRef] [Green Version]
- Bao, F.; Bai, H.Y.; Wu, Z.R.; Yang, Z.G. Phenolic compounds from cultivated Glycyrrhiza uralensis and their PD-1/PD-L1 inhibitory activities. Nat. Prod. Res. 2019, 1–8. [Google Scholar] [CrossRef]
- Han, Y.; Gao, Y.; He, T.; Wang, D.; Guo, N.; Zhang, X.; Chen, S.; Wang, H. PD-1/PD-L1 inhibitor screening of caffeoylquinic acid compounds using surface plasmon resonance spectroscopy. Anal. Biochem. 2018, 547, 52–56. [Google Scholar] [CrossRef]
- Okuda, T.; Yoshida, T.; Mori, K. [Consitutents of Geranium thunbergii Sieb. et Zucc. II. Ellagitannins. (1) (author’s transl)]. Yakugaku Zasshi 1975, 95, 1462–1466. [Google Scholar] [CrossRef] [Green Version]
- Okuda, T.; Mori, K.; Murakami, R. [Constituents of Geranium thunbergii Sieb. et Zucc. VI. Difference of tannin activity caused by structural differences. (2). Colorimetry with methylene blue (author’s transl)]. Yakugaku Zasshi 1977, 97, 1273–1278. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.J.; Choi, H.J.; Park, M.J.; Lee, J.Y.; Jeong, S.I.; Lee, S.; Kim, K.H.; Joo, M.; Jeong, H.S.; Kim, J.E.; et al. The inhibitory effects of Geranium thunbergii on interferon-gamma- and LPS-induced inflammatory responses are mediated by Nrf2 activation. Int. J. Mol. Med. 2015, 35, 1237–1245. [Google Scholar] [CrossRef] [Green Version]
- Calderon-Montano, J.M.; Burgos-Moron, E.; Perez-Guerrero, C.; Lopez-Lazaro, M. A review on the dietary flavonoid kaempferol. Mini Rev. Med. Chem. 2011, 11, 298–344. [Google Scholar] [CrossRef]
- Wang, J.; Fang, X.; Ge, L.; Cao, F.; Zhao, L.; Wang, Z.; Xiao, W. Antitumor, antioxidant and anti-inflammatory activities of kaempferol and its corresponding glycosides and the enzymatic preparation of kaempferol. PLoS ONE 2018, 13, e0197563. [Google Scholar] [CrossRef]
- Xiufen, W.; Hiramatsu, N.; Matsubara, M. The antioxidative activity of traditional Japanese herbs. Biofactors 2004, 21, 281–284. [Google Scholar] [CrossRef]
- Alonso-Castro, A.J.; Ortiz-Sanchez, E.; Garcia-Regalado, A.; Ruiz, G.; Nunez-Martinez, J.M.; Gonzalez-Sanchez, I.; Quintanar-Jurado, V.; Morales-Sanchez, E.; Dominguez, F.; Lopez-Toledo, G.; et al. Kaempferitrin induces apoptosis via intrinsic pathway in HeLa cells and exerts antitumor effects. J. Ethnopharmacol. 2013, 145, 476–489. [Google Scholar] [CrossRef]
- Li, W.; Kim, T.I.; Kim, J.H.; Chung, H.S. Immune Checkpoint PD-1/PD-L1 CTLA-4/CD80 are Blocked by Rhus verniciflua Stokes and its Active Compounds. Molecules 2019, 24, 4062. [Google Scholar] [CrossRef] [Green Version]
- Xia, A.; Zhang, Y.; Xu, J.; Yin, T.; Lu, X.J. T Cell Dysfunction in Cancer Immunity and Immunotherapy. Front. Immunol. 2019, 10, 1719. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.F.; Hsu, P.N. Cancer immunotherapy by targeting immune checkpoints: Mechanism of T cell dysfunction in cancer immunity and new therapeutic targets. J. Biomed. Sci. 2017, 24, 35. [Google Scholar] [CrossRef]
- Cai, J.; Wang, D.; Zhang, G.; Guo, X. The Role Of PD-1/PD-L1 Axis In Treg Development And Function: Implications For Cancer Immunotherapy. Onco Targets Ther. 2019, 12, 8437–8445. [Google Scholar] [CrossRef] [Green Version]
- Minchom, A.; Thavasu, P.; Ahmad, Z.; Stewart, A.; Georgiou, A.; O’Brien, M.E.R.; Popat, S.; Bhosle, J.; Yap, T.A.; de Bono, J.; et al. A study of PD-L1 expression in KRAS mutant non-small cell lung cancer cell lines exposed to relevant targeted treatments. PLoS ONE 2017, 12, e0186106. [Google Scholar] [CrossRef] [Green Version]
- Zhai, W.; Zhou, X.; Du, J.; Gao, Y. In vitro assay for the development of small molecule inhibitors targeting PD-1/PD-L1. Methods Enzymol. 2019, 629, 361–381. [Google Scholar]
- Boohaker, R.J.; Sambandam, V.; Segura, I.; Miller, J.; Suto, M.; Xu, B. Rational design and development of a peptide inhibitor for the PD-1/PD-L1 interaction. Cancer Lett. 2018, 434, 11–21. [Google Scholar] [CrossRef]
- Zak, K.M.; Kitel, R.; Przetocka, S.; Golik, P.; Guzik, K.; Musielak, B.; Domling, A.; Dubin, G.; Holak, T.A. Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1. Structure 2015, 23, 2341–2348. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, A.; Ahmed, M.; Okoye, I.; Arutyunova, E.; Babu, D.; Turnbull, W.L.; Kundu, J.K.; Shields, J.; Agopsowicz, K.C.; Xu, L.; et al. Comprehensive in vitro characterization of PD-L1 small molecule inhibitors. Sci. Rep. 2019, 9, 12392. [Google Scholar] [CrossRef]
- Musielak, B.; Kocik, J.; Skalniak, L.; Magiera-Mularz, K.; Sala, D.; Czub, M.; Stec, M.; Siedlar, M.; Holak, T.A.; Plewka, J. CA-170—A Potent Small-Molecule PD-L1 Inhibitor or Not? Molecules 2019, 24, 2804. [Google Scholar] [CrossRef] [Green Version]
- Sung, Y.Y.; Yoon, T.; Yang, W.K.; Kim, S.J.; Kim, H.K. Anti-obesity effects of Geranium thunbergii extract via improvement of lipid metabolism in high-fat diet-induced obese mice. Mol. Med. Rep. 2011, 4, 1107–1113. [Google Scholar] [CrossRef]
- Mazewski, C.; Kim, M.S.; Gonzalez de Mejia, E. Anthocyanins, delphinidin-3-O-glucoside and cyanidin-3-O-glucoside, inhibit immune checkpoints in human colorectal cancer cells in vitro and in silico. Sci. Rep. 2019, 9, 11560. [Google Scholar] [CrossRef] [Green Version]
- Dabeek, W.M.; Marra, M.V. Dietary Quercetin and Kaempferol: Bioavailability and Potential Cardiovascular-Related Bioactivity in Humans. Nutrients 2019, 11, 2288. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.W.; Zhang, J.Y.; Xu, W.; Li, J.; Zhang, W.Q. [The biotransformation of kaempferitrin by human intestinal flora]. Yao Xue Xue Bao 2005, 40, 717–721. [Google Scholar]
- DuPont, M.S.; Day, A.J.; Bennett, R.N.; Mellon, F.A.; Kroon, P.A. Absorption of kaempferol from endive, a source of kaempferol-3-glucuronide, in humans. Eur. J. Clin. Nutr. 2004, 58, 947–954. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Wang, M.; Guo, Z.; Zhang, X. Pharmacokinetic evaluation of the interaction between oral kaempferol and ethanol in rats. Acta Pharm. 2016, 66, 563–568. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.G.; Kim, Y.S.; Kim, J.H.; Chung, H.S. Antiviral activity of ethanol extract of Geranii Herba and its components against influenza viruses via neuraminidase inhibition. Sci. Rep. 2019, 9, 12132. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]





| Protein | KD (M) | ka (M−1 s−1) | kd (s−1) | R2 |
|---|---|---|---|---|
| PD-1 | 3.11 × 10−5 | 3.15 × 102 | 9.79 × 10−3 | 0.9925 |
| PD-L1 | 1.97 × 10−5 | 3.92 × 102 | 7.73 × 10−3 | 0.9958 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Kim, Y.S.; Choi, J.-G.; Li, W.; Lee, E.J.; Park, J.-W.; Song, J.; Chung, H.-S. Kaempferol and Its Glycoside, Kaempferol 7-O-rhamnoside, Inhibit PD-1/PD-L1 Interaction In Vitro. Int. J. Mol. Sci. 2020, 21, 3239. https://doi.org/10.3390/ijms21093239
Kim JH, Kim YS, Choi J-G, Li W, Lee EJ, Park J-W, Song J, Chung H-S. Kaempferol and Its Glycoside, Kaempferol 7-O-rhamnoside, Inhibit PD-1/PD-L1 Interaction In Vitro. International Journal of Molecular Sciences. 2020; 21(9):3239. https://doi.org/10.3390/ijms21093239
Chicago/Turabian StyleKim, Ji Hye, Young Soo Kim, Jang-Gi Choi, Wei Li, Eun Jin Lee, Jin-Wan Park, Jaeyoung Song, and Hwan-Suck Chung. 2020. "Kaempferol and Its Glycoside, Kaempferol 7-O-rhamnoside, Inhibit PD-1/PD-L1 Interaction In Vitro" International Journal of Molecular Sciences 21, no. 9: 3239. https://doi.org/10.3390/ijms21093239