1. Introduction
Toll-like receptor 3 (TLR3) is an important pattern recognition receptor that recognizes double-stranded RNA (dsRNA), which is a viral replication intermediate [
1], and its synthetic analog polyinosinic:polycytidylic acid [
2]. TLR3 is predominantly expressed in the endosomal compartment of sentinel cells such as macrophages and myeloid dendritic cells, where the recognition of endocytosed dsRNA occurs at an acidic pH (pH ≤ 6.5) [
3,
4]. The activated TLR3 mounts a strong immune response against invading viruses by triggering the expression of proinflammatory cytokines, typically antiviral interferons (IFNs) [
5]. The agonist-mediated conformational change in the receptor initiates the recruitment of an adaptor, Toll/interleukin-1 receptor (TIR) domain-containing adaptor protein inducing IFN-β (TRIF). Subsequent stages of the signaling cascade involve the recruitment of the inhibitor of nuclear factor-κB-kinase-ε (IKK-ε) complex and the phosphorylation of TANK-binding kinase 1 (TBK1) [
6]. The phosphorylated TBK1 activates the transcription factor IFN-regulatory factor 3 (IRF3), which induces the expression of type I IFNs (IFN-α/β) [
7]. In addition, dsRNA-mediated TLR3 signaling leads to the activation of a number of transcription factors, specifically nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), IRF-7, and members of the activator protein 1 (AP-1) family [
8].
TLR3 exhibits a tripartite domain architecture, comprising a leucine-rich repeat (LRR)-rich ectodomain (ECD), a transmembrane (TM) domain, and a cytosolic Toll/interleukin-1 receptor (TIR) domain [
9]. The structure of mouse TLR3-ECD complexed with dsRNA has been resolved through X-ray crystallography [
10], and the nuclear magnetic resonance (NMR) structure of the TM domain in its dimeric and trimeric forms have been reported [
11]. However, the structure of the TIR domain or the full-length, intact TLR3 structure has remained elusive. Phylogenetic analysis has categorized vertebrate TLRs into different subfamilies, including TLR1/2/6/10, TLR3, TLR4, TLR5, TLR7/8/9, and TLR11/12/13/21/22/23, suggesting that TLR3 belongs to a unique category within the TLR family [
12]. Because TLR3 triggers a strong immune response against viral infection, it is considered a promising drug target for the development of effective vaccine adjuvants [
13]. The dsRNA-specificity of TLR3 is independent of nucleotide composition; therefore, a large number of potent adjuvants has been developed and investigated in clinical trials [
14,
15]. Although TLR3 signaling is crucial for the protection of the host against viral infections [
16], dysregulated signaling has been implicated in several inflammatory disorders [
17], autoimmune diseases [
18], cancer [
19], and atherosclerosis [
20]. Synthetic monoclonal antibodies [
21,
22] and low-molecular-weight compounds [
23,
24] have shown beneficial effects on TLR3-mediated diseases in mice and human subjects [
25].
Despite the clinical significance of TLR3 modulation, the structural organization of full-length TLR3 in an activated state has not been experimentally determined, probably due to its complex architecture that spans the membrane and the aqueous environment of the cell. Earlier, Liu et al. (2008) have proposed a full-length model of TLR3 [
10]; however, the atomistic details of inter-domain interactions, time-dependent structural evolution, and dynamics of TM, TIR, and ECD with respect to the phospholipid bilayer as a single unit have not been thoroughly studied. In the present study, we analyzed a full-length homodimerization complex of TLR3 embedded in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer using molecular dynamics (MD) simulation techniques. The membrane-solvated TLR3 model was able to elucidate several important structural features of the individual ECD, TM, and TIR domains in a physiologically applicable environment. The ECD and TIR domains are established drug targets; therefore, a detailed knowledge about their structural topology and intermolecular interactions is crucial for the development of effective, novel adjuvant, or antagonistic candidates.
3. Discussion
TLR activation consists of a complex assembly of multimeric receptor and adaptor molecules in the cell or endosomal membrane [
40]. Of these, ligand-induced homo/heterodimeric receptors are considered the basic signaling unit required for signal transduction [
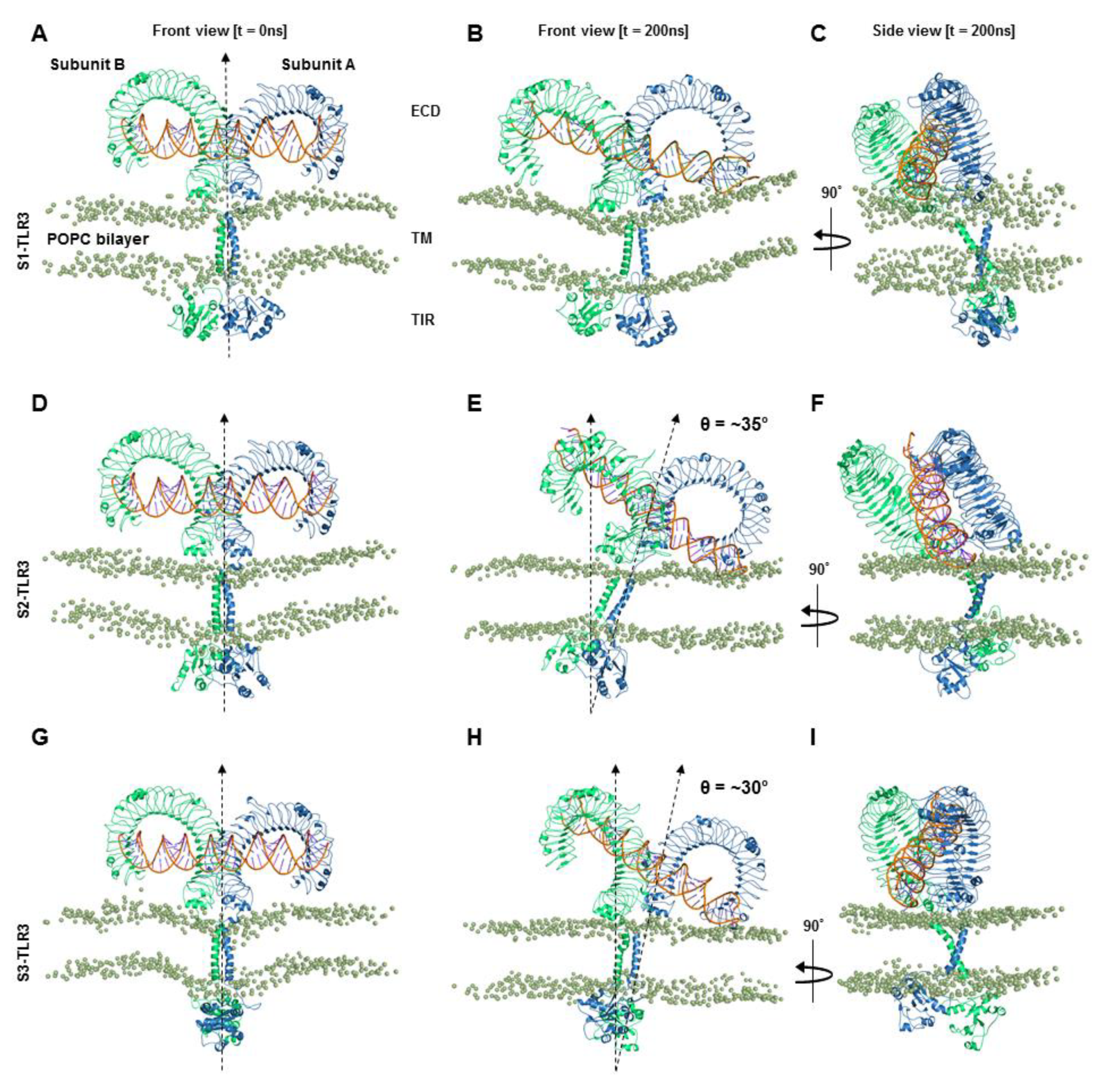
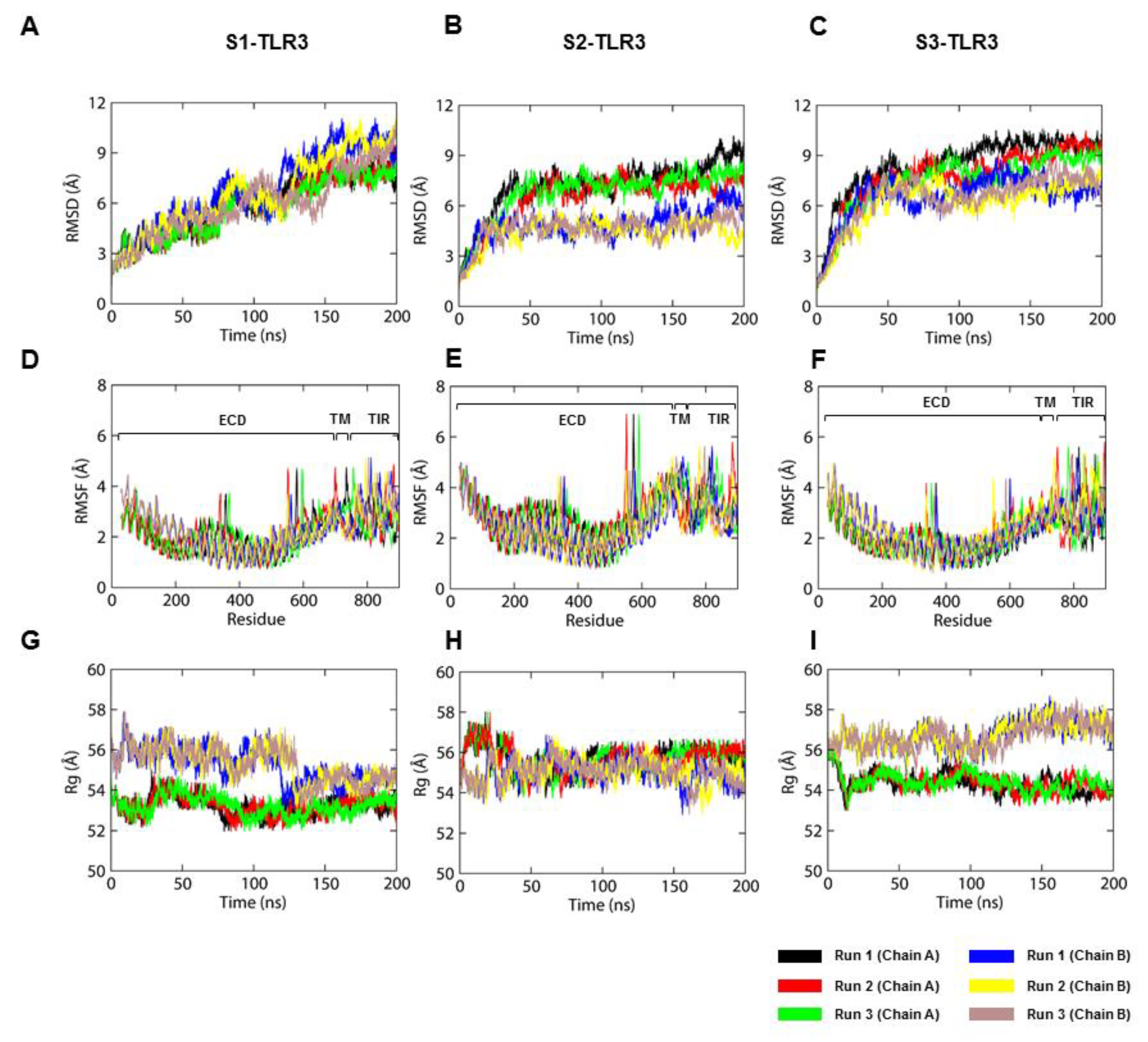
9]. Here, we studied three membrane-solvated models of 2:1 TLR3-dsRNA homodimerization complexes and identified the model most likely to represent the signaling-competent receptor in a physiological environment. The TIR domain of each full-length receptor was constructed via homology modeling using the crystal structures of either the TLR2-, TLR6-, or TLR10-TIR homodimer as templates. A comparative analysis of three MD trajectories suggested that the TLR3-TIR homodimer built from the TLR6-TIR domain led to a full-length receptor with the stability necessary to maintain key intermolecular interactions with the ligand and with the membrane, as exhibited by the native form.
Our simulation data indicated that the long, flexible juxtamembrane loops of TLR3 allow for the simultaneous bending of the ECD and TIR domains on both surfaces of the membrane. The cytoplasmic linker connecting the TM and TIR domains has been reported to be essential for the subcellular targeting of TLR3 toward the endosomal membrane [
41]. The juxtamembrane regions are also believed to play an important role in the membrane-anchoring mechanisms of TLR4 [
29,
42,
43,
44]. A previous study of the chimeric TLR3-TLR9 receptor has demonstrated that TLR3 can also be localized to the plasma membrane by means of its ECD but that endocytosis of the receptor-ligand complex and endosomal acidification are required for efficient signaling [
45]. We observed that the membrane-anchored TLR3 progressively tilts on the bilayer surface due to the electrostatic attraction between the charged microdomains of both the protein and phospholipids. Although the ECD exhibited a sharp tilt on the membrane, the LRR-NT was partially absorbed by the lipid headgroups; in contrast, the LRR-NT of our previously reported TLR4 model was completely buried in the bilayer surface [
29]. It is possible that the negatively charged dsRNA that spans the entire length of the TLR3-ECD restricted the insertion of LRR-NT into the membrane surface, owing to the electrostatic repulsion of the phospholipid headgroups. The role of LRR-NT in the tilting and membrane anchoring of TLR3 seems to agree with the lateral clustering model of TLR3, where multiple ECDs cluster on their lateral surfaces to recognize dsRNA molecules that are up to 90 base pairs long [
46]. The convex surface spanning LRRs 3–7, which is distinct from the N-terminal dsRNA-binding site (i.e., LRR-NT and LRRs 1–3), mediates the lateral clustering of ECDs. This indicates that tilted ECDs may facilitate the lateral clustering of TLR3 even though LRR-NT is partially buried in the membrane during ligand-induced signal transduction.
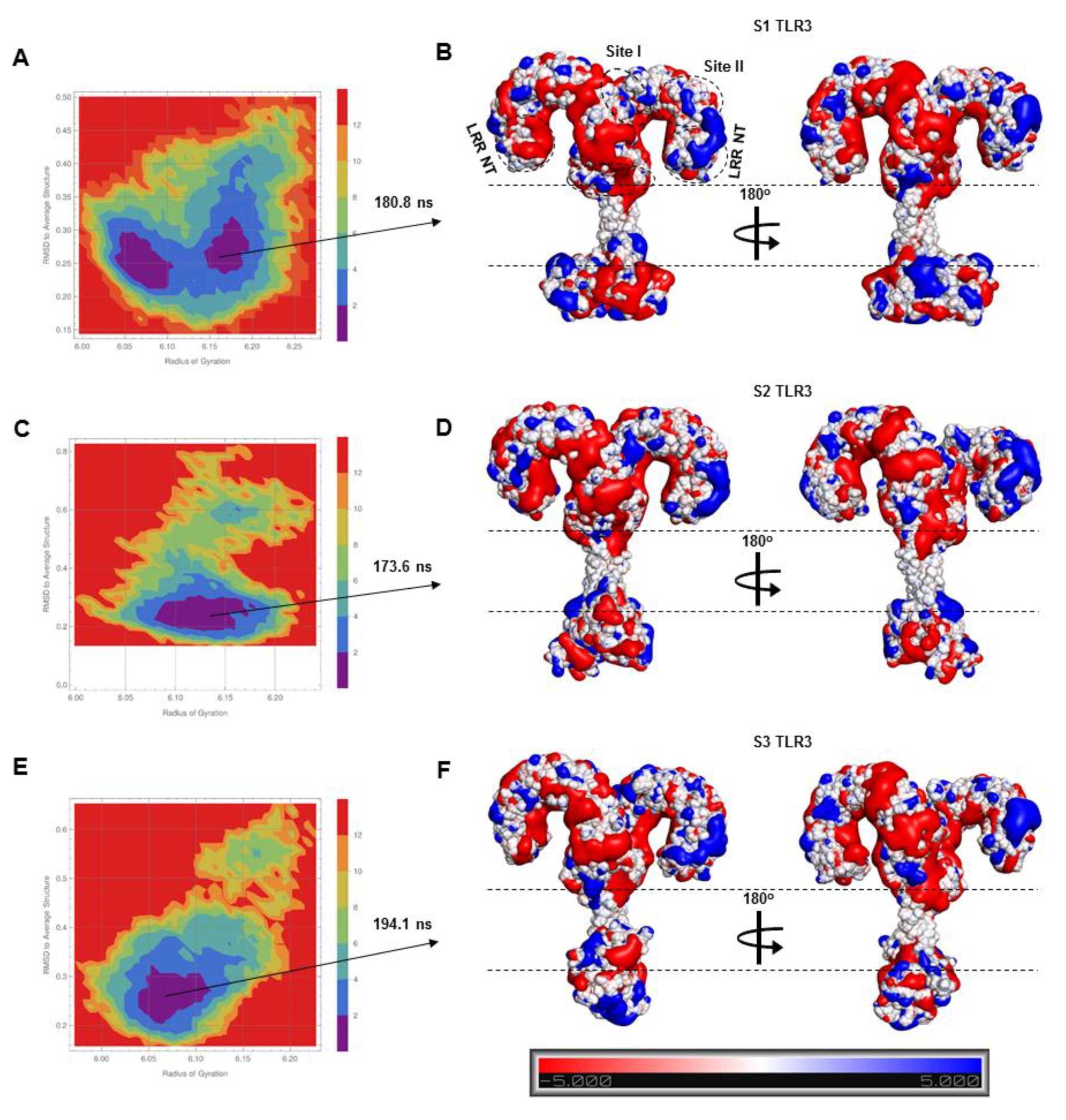
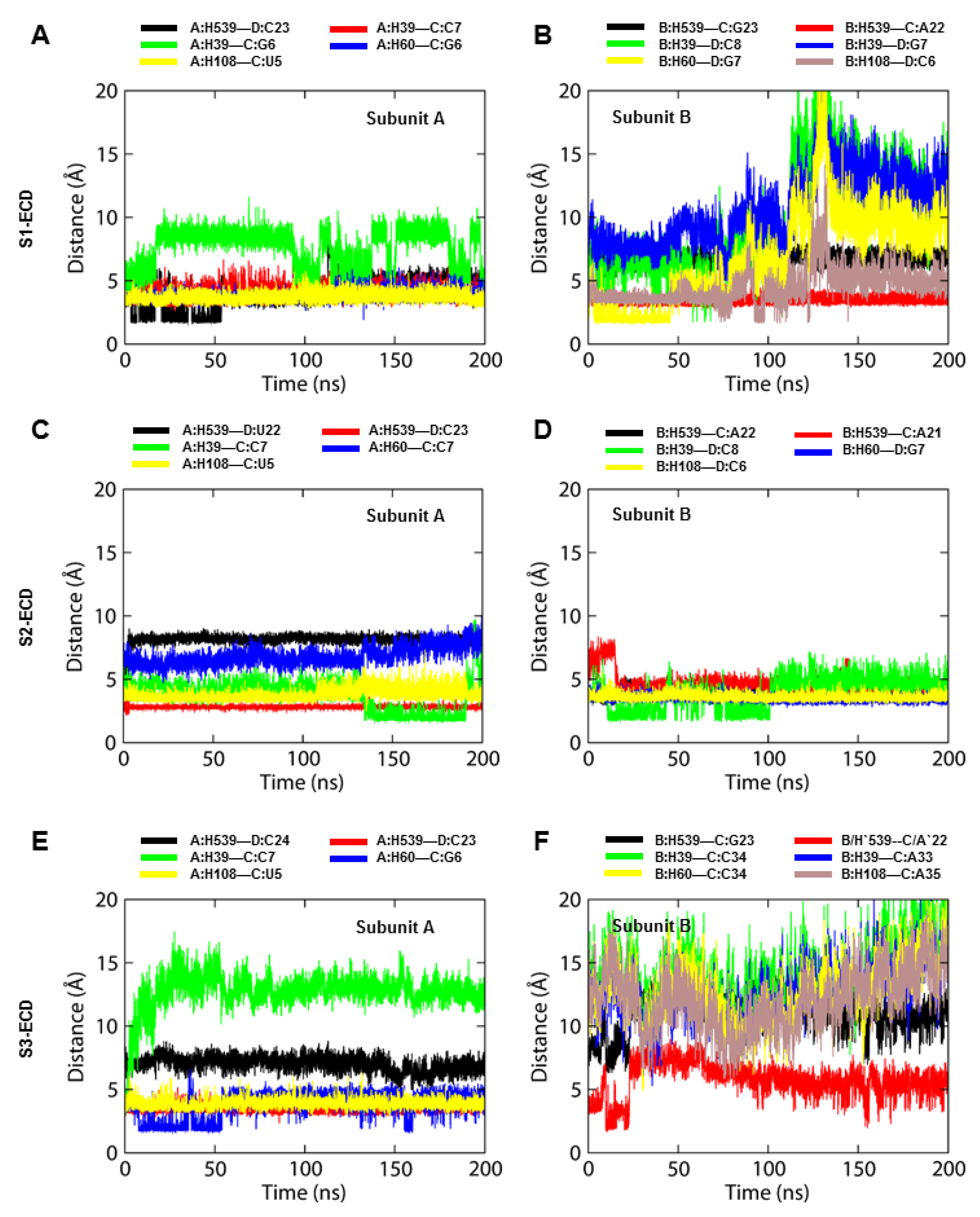
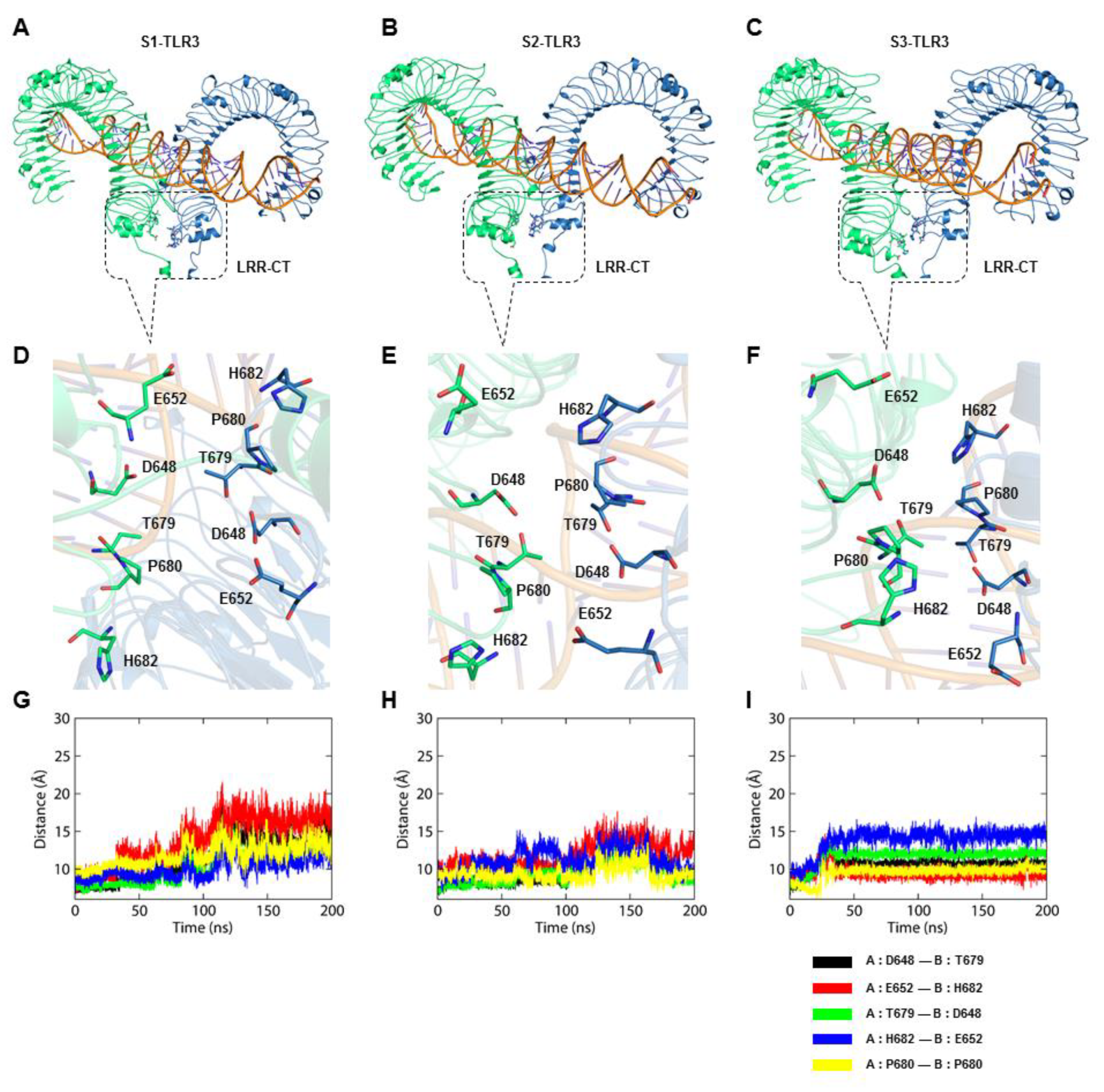
The bound dsRNA retained its structural integrity through the stable interaction with TLR3, indicating that the dynamic behavior of the ECD does not significantly alter the dsRNA binding pattern. The X-ray crystal structure of mouse TLR3-ECD (PDB ID: 3CIY) demonstrated that dsRNA forms two major contacts with the glycan-free surfaces of the ECD: one at the C-terminal site (site I) and the other at the N-terminal site (site II). Site I comprises the conserved residues N515, N517, H539, N541, and R544, while site II consists of residues H39, H60, R64, F84, T86, H108, and E110. The mutations H539E and N541A at site I and the mutations H39A and H60A at site II abrogated the TLR3 response towards dsRNA, indicating these residues are important for ligand recognition [
10]. The most critical electrostatic interaction occurs between the imidazole rings of histidine residues and the phosphate groups of the dsRNA backbone [
9]. The S2-TLR3 model exhibited a stable interaction between H39/H60 and the dsRNA backbone over the course of the simulation. Although the site II residues of S2-TLR3 formed several transient H-bonds and salt bridges with the phosphate groups of dsRNA, the electrostatic and van der Waals forces were crucial for the stronger binding affinity of the receptor–ligand complex. The variable H-bond/salt bridges at site II may be due to the progressive tilt of the ECD and the slightly twisted dsRNA backbone during the simulation. Previous studies have shown that dsRNA binds to a preformed TLR3 homodimer stabilized by interactions between the LRR-CT domains and that an intact dimerization site is required for both ligand binding and downstream signaling [
47,
48]. The homodimerization site consists of two H-bond pairs, D648/T679 and G652/H682, and the structural proline P680. Mutational analysis revealed that D648A, T679A, and P680L variants are inactive against dsRNA whereas H682A and E652A are partially active [
33]. Our analysis revealed that S2-TLR3 displays an intact homodimerization interface supported by stable spatial contacts among D648/T679 and G652/H682 residue pairs. This result suggests that the equilibrated model of full-length S2-TLR3 embedded in the phospholipid bilayer might represent the physiologically relevant state of an activated receptor.
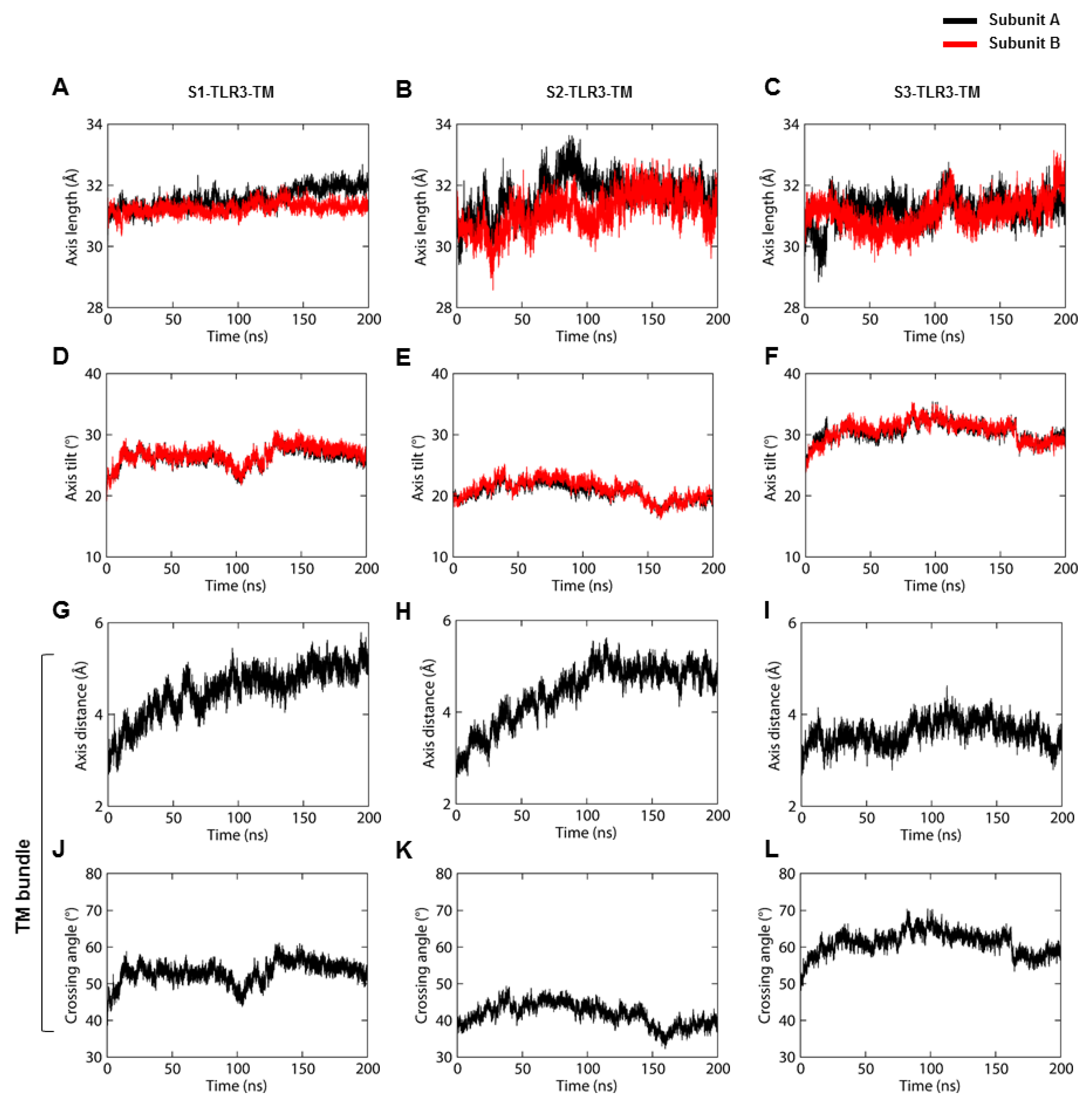
The tilted architecture of the ECD is followed by the tilting and curvature of the TM helices, causing severe topological changes in the TIR domains. The minor distortion of TM helices is a regular phenomenon that occurs due to a hydrophobic mismatch with the bilayer core [
43]. However, the helical transformation of TLR3 was largely distinct from the TLR4-TM previously reported [
29,
42]. This indicates that the orientation and conformational changes of each TLR type may vary, depending on their location in the membrane or the lipid composition of that particular area. Membrane proteins usually exist within specialized microdomains called lipid rafts, which contain different arrangements of localized lipids [
49]. Therefore, the liquid order of a particular membrane microdomain might have an impact on the orientation behavior of membrane proteins, as shown here [
50]. It has been reported that the NMR structure of an isolated TLR3-TM domain in dodecylphosphocholine micelles exists as a right-handed dimer with a helix-crossing angle of –51.1° [
51]. The substantial tilt and curvature and the consistent crossing angle of TM helices in our simulations indicate that the hydrophobic mismatch shapes the TM domain organization of full-length TLR3 in the cell membrane [
52]. As in the full-length structure of TLR4 [
29], the membrane-spanning segment of TLR3 is a relatively long helix of ~30 residues, which included 21 residues of the TM domain and rest from the juxtamembrane segments. Analysis of the amino acid composition of the TM domains revealed the presence of several aromatic residues and relatively polar residues. We observed a cluster of aromatic and aliphatic residues that were entangled in the aromatic stacking and hydrophobic contacts at the C-terminal end of the TM bundle. Partial charges on aromatic side chains can also interact with other aromatic or polar side chains through electrostatic interaction [
53]. We also found that residues M707 and I722 from either end of the TM helix could contribute to the dimerization surface through their long, branched aliphatic side chains. The isolated TLR3-TM domains have the ability to form homodimers or trimers, stabilized by aromatic stacking and van der Waals interaction between the bulky side chains [
11]. While both the dimeric and trimeric forms of TM domain are possible in a micellar environment, the oligomer form of the entire TLR3 molecule in a biological membrane (such as endosomal or lysosomal membranes) needs further experimental clarification.
Ligand recognition enables conformational changes in the TLR structure, thus bringing the TIR domains into close proximity [
54]. Because the experimental structure of the TLR3-TIR domain is unknown, computational models have been used to understand the structure, dynamics, and molecular basis of TLR3-mediated signal transduction [
55,
56]. Based on the sequence homology, the crystal structure of the TLR10-TIR domain seems to be the most suitable template for modeling TLR3-TIR structure (
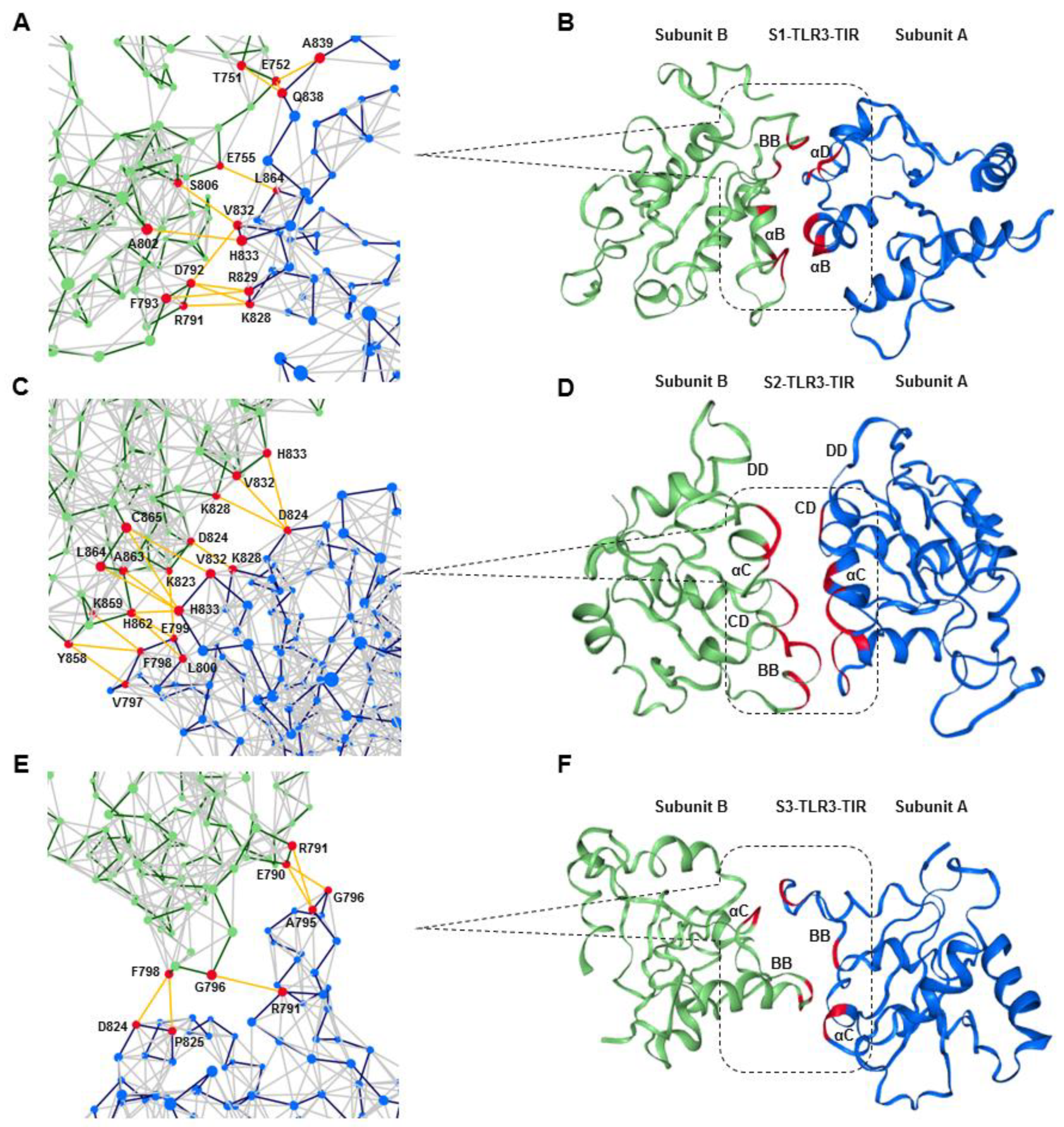
Supplementary material, Figure S6). Although the prediction of dimeric TLR3-TIR architecture is challenging, especially in the absence of biochemical or mutagenesis data, we used multiple homology models followed by MD optimizations to address such problem. Our results showed that the TIR domain architecture of TLR2 or TLR10 was not compatible with the full-length TLR3 in the phospholipid bilayer because the interaction between monomers under dynamic condition was inconsistent or relatively less stable. On the other hand, the architecture of the TLR6-TIR dimer was remarkably stable with an appropriate packing interaction between monomers in the dynamic condition. The main dimerization interface in the TLR6-TIR crystal structure is defined by reciprocal contact between the residues from the CD loop, DD loop, and αC helix of both monomers [
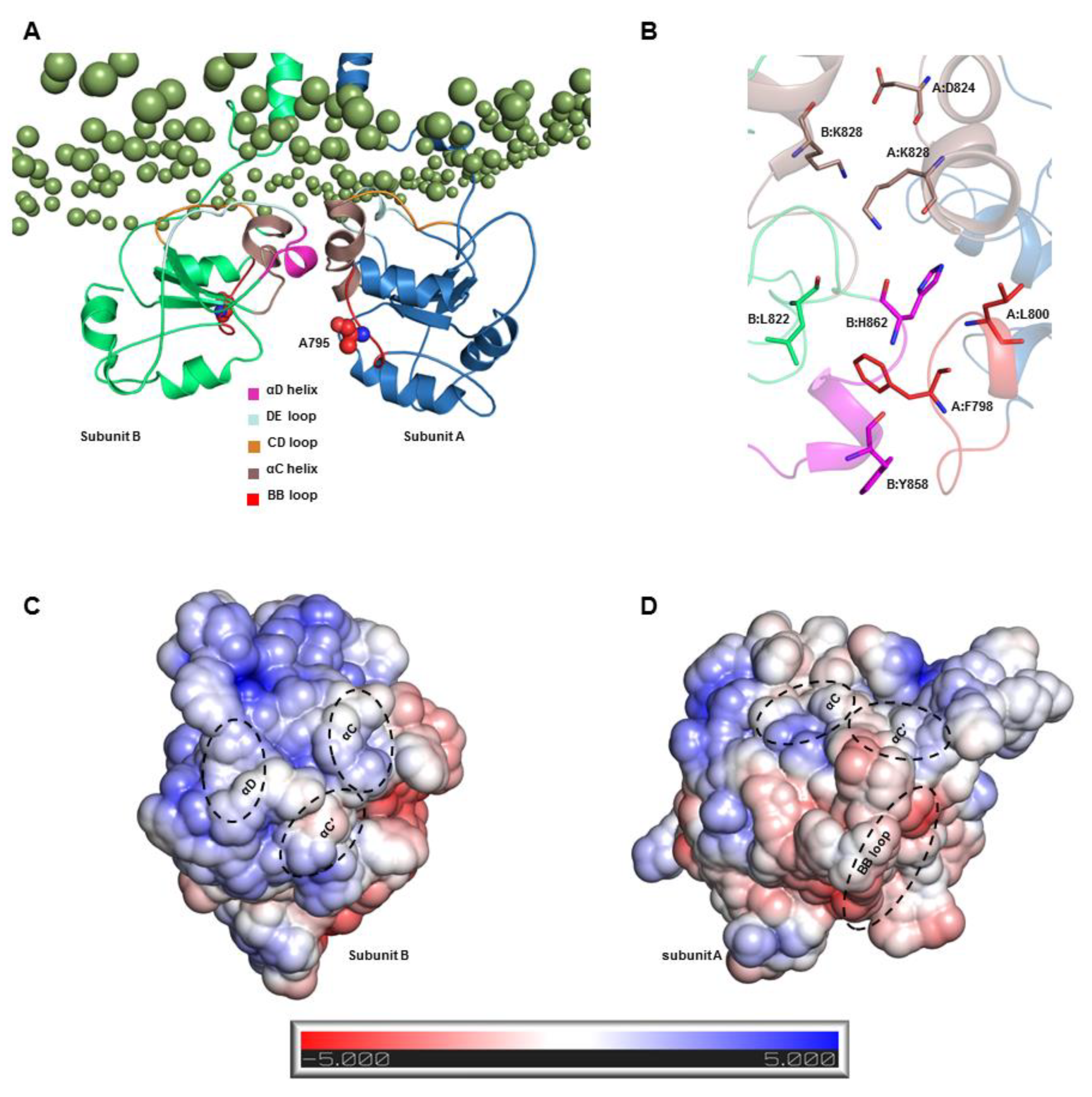
27]. Our equilibrated S2-TLR3 structure exhibited a TIR-TIR interface involving residues from the αC and αD helix and the CD and DE loops of both monomers. The BB loop of one subunit was completely solvent-exposed, while that of the other was partially involved in dimer packing. The BB loop of several TLRs contains the conserved RDXXP motif, which is critical for receptor oligomerization and adaptor recruitment [
54]. In TLR3, the proline of the RDXXP is replaced by an alanine (A795), which has been found to be essential for the recruitment of the TRIF adaptor [
37]. Our analysis of the S2 MD trajectory revealed that A795 is excluded from the homodimerization interface over the course of the entire simulation, suggesting that the BB loop together with A795 can be readily available for TRIF binding. Thus, we assume that the S2-TLR3 model better explains the signaling-competent form of the full-length receptor bound to an agonist. Previous reports have demonstrated that the sequential phosphorylation of Y858 and Y759 is required to initiate TLR3 signaling for the activation of IRF3 and NF-κB [
57,
58,
59]. We found that the side chain of Y858 (αD helix) face opposite to F798 (BB loop) and its hydroxyl group is exposed to the solvent. Likewise, the hydroxyl group of Y759 (βA strand) faced the surface, increasing the likelihood of a phosphorylation reaction. This suggests that the architecture of the S2-TIR domain in full-length TLR3 agrees with current biochemical and mutagenesis data regarding TLR3 signal transduction.
In conclusion, we modeled the structural organization of full-length TLR3 and identified key molecular interactions that are critical for receptor activation under physiological conditions. The ECD of TLR3 is able to orient towards either direction of the membrane surface, while TM helices tilt with a consistent crossing angle to overcome the hydrophobic mismatch. The upper surfaces of the TIR domain were partially absorbed into the lower surface of the membrane bilayer, while the dimer interface comprises the αC, αD helix, and the CD and DE loops from both monomers in an equilibrated TLR3 receptor. The BB loop of one subunit contributes to the dimer packing, while the other subunit remained solvent-exposed throughout the MD simulation, confirming the importance of this segment in adaptor recruitment by the activated receptor (
Supplementary Materials, Model S1). Collectively, our MD simulation data provides crucial insights into the signaling-competent form of the full-length TLR3-dsRNA complex that could be used to inform the future design of therapeutic peptide or small molecule drug candidates.