Chlorinated Auxins—How Does Arabidopsis Thaliana Deal with Them?
Abstract
:1. Introduction
2. Results
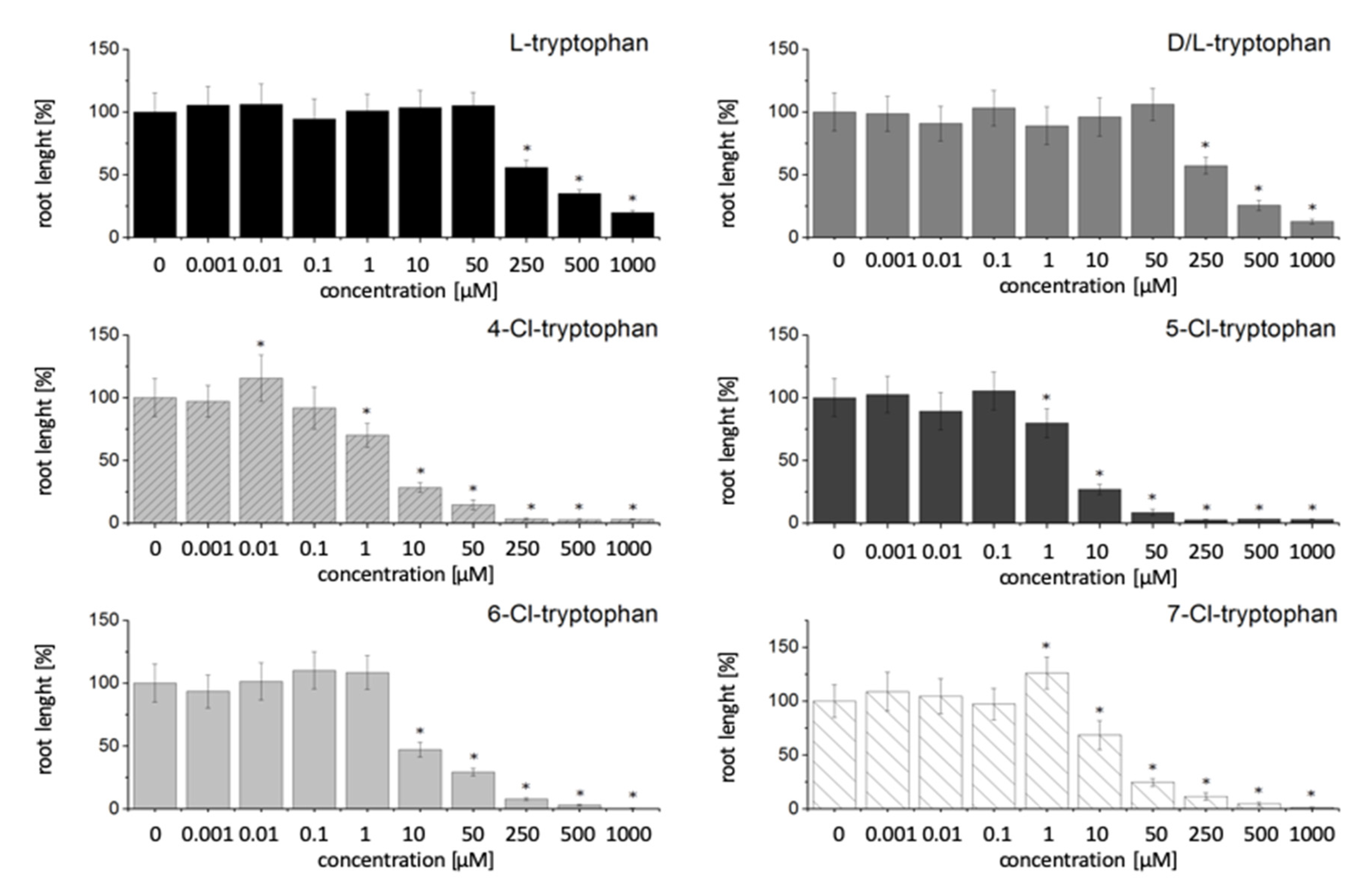
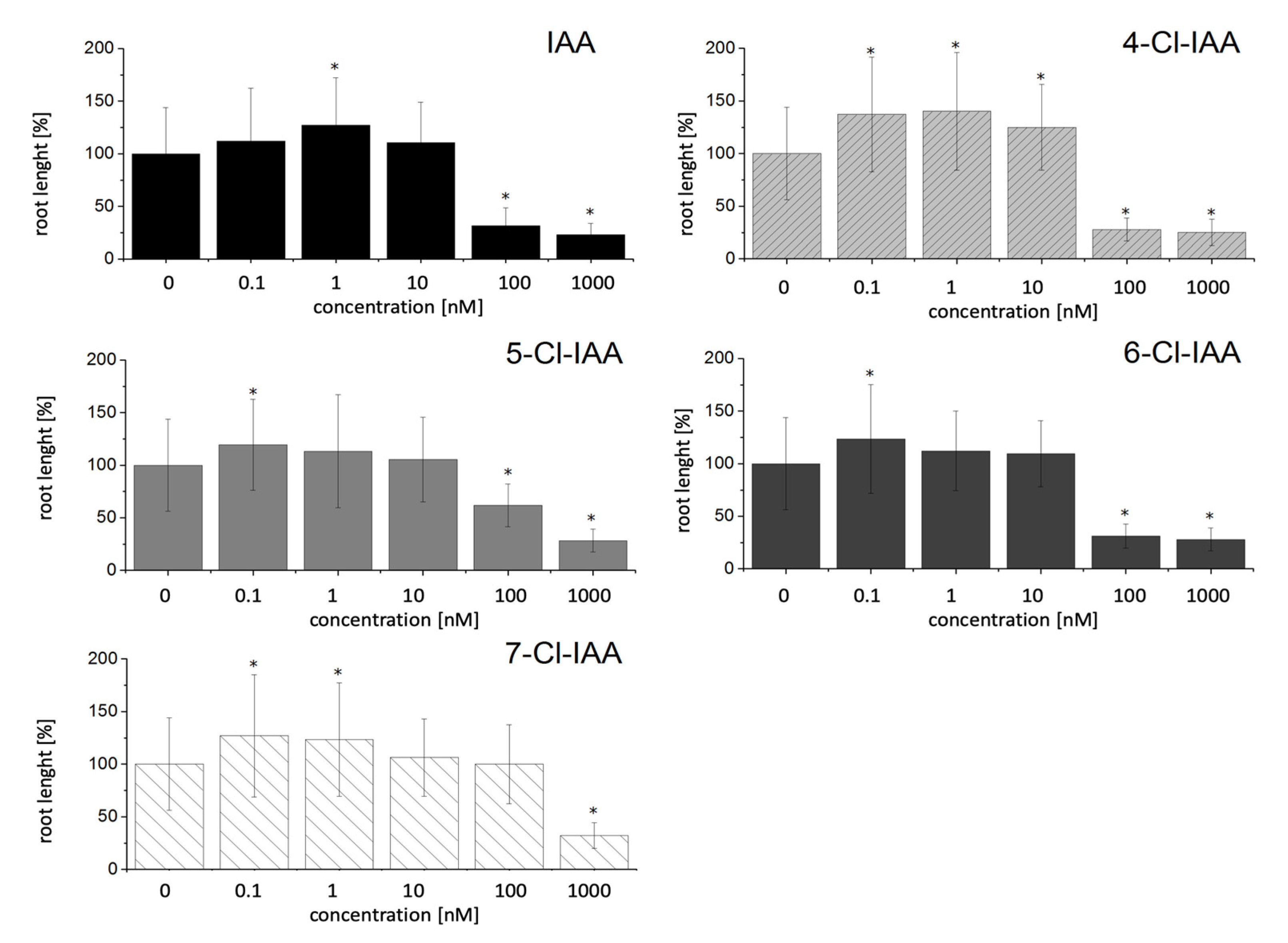
2.1. Effect of Halogenated Compounds on the Growth of Arabidopsis thaliana Wild Type
2.2. Chlorinated Compounds Produced in A. thaliana Wild Type after Incubation with Chlorinated Trp Derivatives
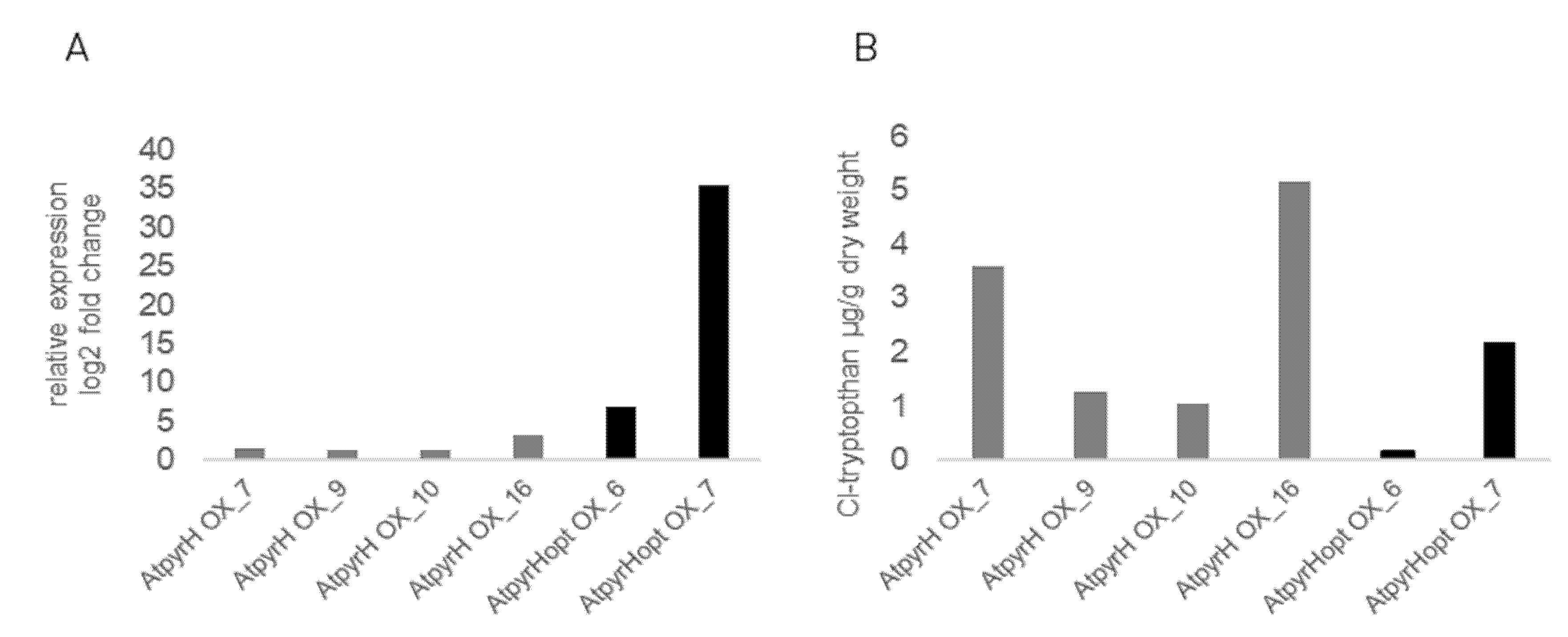
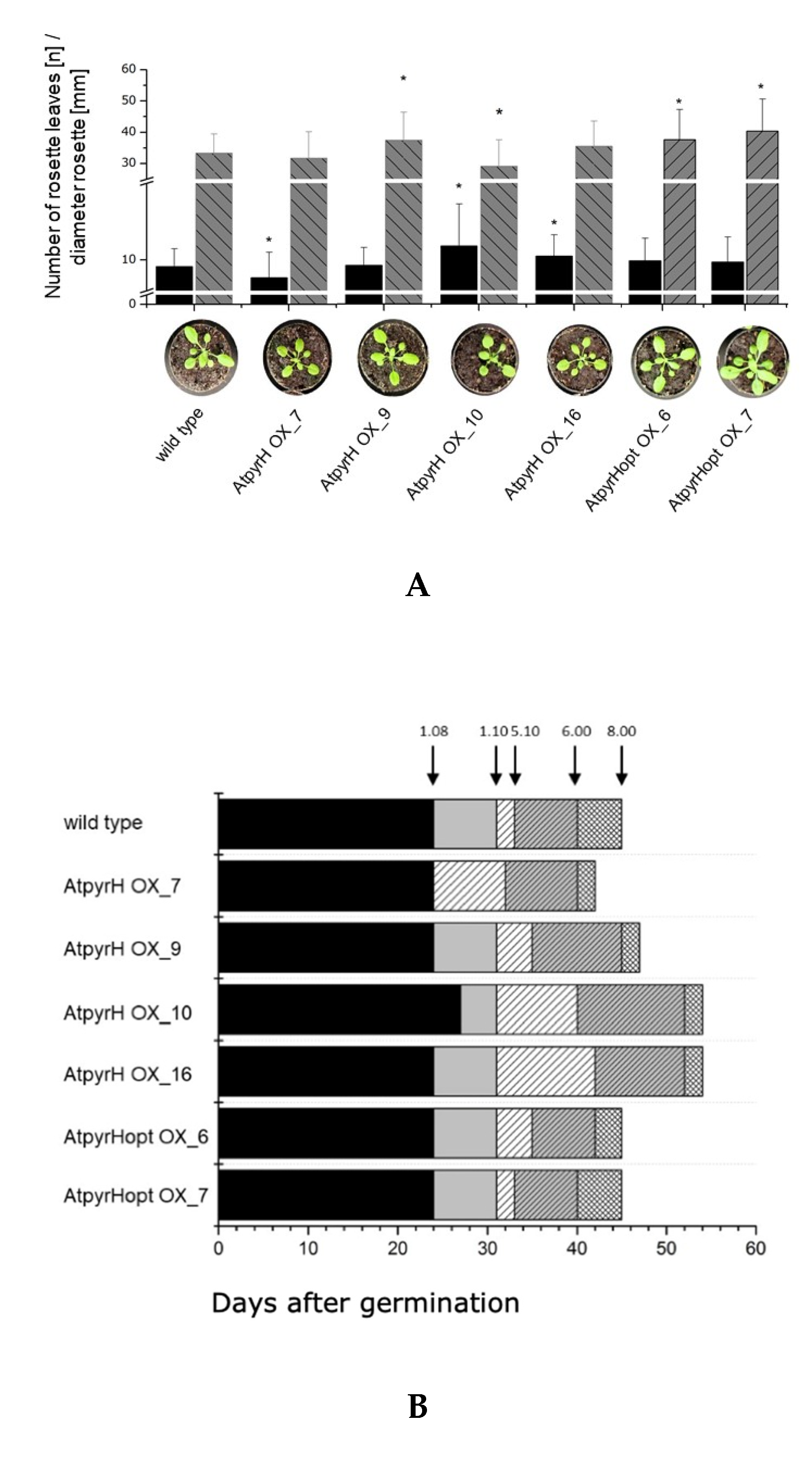
2.3. Chlorinated Compounds Are Produced in Transgenic A. thaliana Lines but Do Not Alter Growth
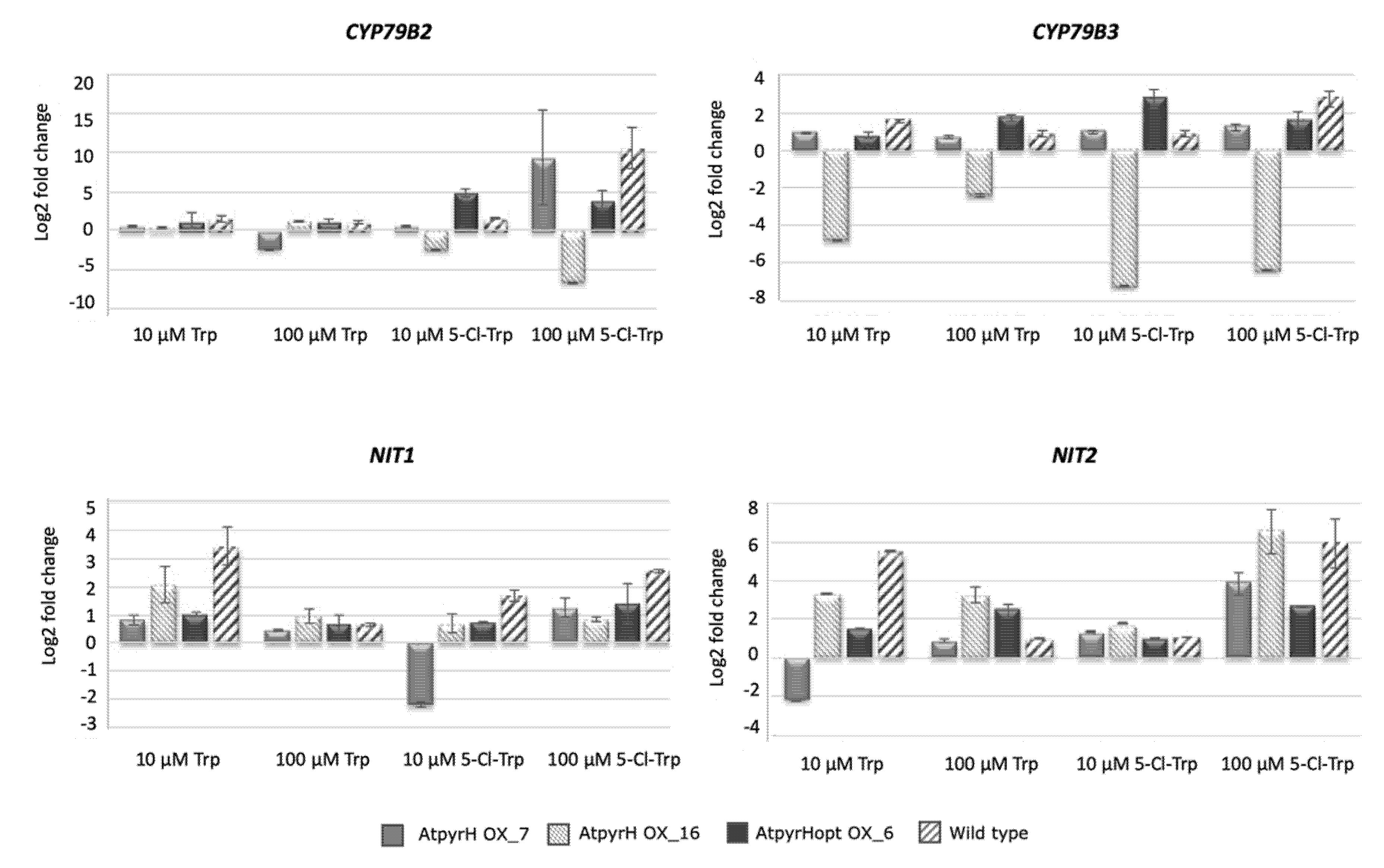
2.4. Analysis of Gene Expression from the Trp-Dependent IAA Biosynthetic Pathway in Different Transgenic A. thaliana Lines
3. Discussion
3.1. Effect of Halogenated Compounds with an Indole Moiety on Growth of A. thaliana Wild Type
3.2. Which Chlorinated Compounds Can Be Produced in A. thaliana Wild Type?
3.3. Chlorinated Compounds in A. thaliana and Their Effect on Plant Growth
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. RNA Extraction and qRT-PC Analysis
4.3. UPLC-QqQ-MS Analysis
4.4. Enzyme Assay for GH3 Activity and TLC Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 4-Cl-IAA | 4-chloro-indole-3-acetic acid |
| 5-Cl-IAA | 5-chloro-indole-3-acetic acid |
| 6-Cl-IAA | 6-chloro-indole-3-acetic acid |
| 7-Cl-IAA | 7-chloro-indole-3-acetic acid |
| 4-Cl-IAN | 4-chloro-indole-3-acetonitrile |
| 5-Cl-IAN | 5-chloro-indole-3-acetonitrile |
| 6-Cl-IAN | 6-chloro-indole-3-acetonitrile |
| 7-Cl-IAN | 7-chloro-indole-3-acetonitrile |
| 4-Cl-Trp | 4-chloro-indole -tryptophan |
| 5-Cl-Trp | 5-chloro-indole -tryptophan |
| 6-Cl-Trp | 6-chloro-indole -tryptophan |
| 7-Cl-Trp | 7-chloro-indole -tryptophan |
| 5-F-IAA | 5-fluoro-indole-3-acetic acid |
| CYP79B2 | cytochrome P450 79B2 |
| CYP79B3 | cytochrome P450 79B3 |
| EB | Ethidium bromide |
| GH3 | Gretchen Hagen 3 enzyme |
| GST | Glutathione-S transferase |
| IAA | indole-3-acetic acid |
| IAA-Asp | IAA conjugate with aspartate |
| IAA-Trp | IAA conjugate with tryptophan |
| IAN | Indole-3-acetonitrile |
| IBA | indole-3-butyric acid |
| IPTG | isopropyl-β-d-thiogalactopyranoside |
| LB | Luria Bertani medium |
| MS | Murashige & Skoog medium |
| NIT1 | nitrilase 1 |
| NIT2 | nitrilase 2 |
| TLC | thin layer chromatography |
| Trp | tryptophan |
References
- Reinecke, D.M. 4-Chloroindole-3-acetic acid and plant growth. J. Plant Growth Regul. 1999, 27, 3–13. [Google Scholar] [CrossRef]
- Rescher, U.; Walther, A.; Schiebl, C.; Klämbt, D. Binding Affinities of 4-Chloro-, 2-Methyl-, 4-Methyl-, and 4-Ethylindoleacetic Acid to Auxin-binding Protein 1 (ABP1) Correlate with Their Growth-Stimulating Activities. J. Plant Growth Regul. 1996, 15, 1–3. [Google Scholar] [CrossRef]
- Karcz, W.; Burdach, Z. A comparison of the effects of IAA and 4-Cl-IAA on growth, proton secretion and membrane potential in maize coleoptile segments. J. Exp. Bot. 2002, 53, 1089–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marumo, S.; Hattori, H.; Yamamoto, A. Plant Growth Substances; Company Hirokawa Publishing: Tokyo, Japan, 1973; pp. 419–428. [Google Scholar]
- Ozga, J.A.; Reinecke, D.M.; Ayele, B.T.; Ngo, P.; Nadeau, C.; Wickramarathna, A.D. Developmental and Hormonal Regulation of Gibberellin Biosynthesis and Catabolism in Pea Fruit. Plant Physiol. 2009, 150, 448–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, M.G.; Reinecke, D.M.; Ozga, J.A. The Auxins IAA and 4-Cl-IAA Differentially Modify Gibberellin Action via Ethylene Response in Developing Pea Fruit. J. Plant Growth Regul. 2005, 24, 214–225. [Google Scholar] [CrossRef]
- Gandar, J.-C.; Nitsch, C. Isolement de l’ester méthylique d’un acide chloro-3-indolylacétique à partir de graines immatures de Pois Pisum sativum. Comptes Rendus Hebd. Séances l’Académie Sci. 1967, 265, 1795–1798. [Google Scholar]
- Marumo, S.; Hattori, H.; Abe, H.; Munakata, K. Isolation of 4-Chloroindolyl-3-acetic Acid from Immature Seeds of Pisum sativum. Nature 1968, 219, 959–960. [Google Scholar] [CrossRef]
- Marumo, S.; Abe, H.; Hattori, H.; Munakata, K. Isolation of a Novel Auxin, Methyl 4-Chloroindoleacetate from Immature Seeds of Pisum sativum. Agric. Biol. Chem. 1986, 32, 117–118. [Google Scholar] [CrossRef]
- Schneider, E.A.; Kazakoff, C.W.; Wightman, F. Gas chromatography-mass spectrometry evidence for several endogenous auxins in pea seedling organs. Planta 1985, 165, 232–241. [Google Scholar] [CrossRef]
- Engvild, K.C. Natural Chlorinated Auxins Labelled with Radioactive Chloride in Immature Seeds. Physiol. Plant. 1975, 34, 286–287. [Google Scholar] [CrossRef]
- Hofinger, M.; Böttger, M. Identification by GC-MS of 4-Chloroindolylacetic acid and its methyl ester in immature Vicia faba seeds. Phytochemistry 1979, 18, 653–654. [Google Scholar] [CrossRef]
- Katayama, M.; Thiruvikraman, S.V.; Marumo, S. Identification of 4Chloroindole-3-acetic Acid and Its Methyl Ester in Immature Seeds of Vicia amurensis (the Tribe Vicieae), and Their Absence from Three Species of Phaseoleae. Plant Cell Physiol. 1987, 28, 383–386. [Google Scholar] [CrossRef]
- Magnus, V.; Ozga, J.A.; Reinecke, D.M.; Pierson, G.L.; Larue, T.A.; Cohen, J.D.; Brenner, M.L. 4-chloroindole-3-acetic and indole-3-acetic acids in Pisum sativum. Phytochemistry 1997, 46, 675–681. [Google Scholar] [CrossRef]
- Pless, T.; Böttger, M.; Hedden, P.; Graebe, J. Occurrence of 4-Cl-Indoleacetic Acid in Broad Beans and Correlation of Its Levels with Seed Development. Plant Physiol. 1984, 74, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Lam, H.K.; McAdam, S.A.M.; McAdam, E.L.; Ross, J.J. Evidence That Chlorinated Auxin Is Restricted to the Fabaceae But Not to the Fabeae. Plant Physiol. 2015, 168, 798–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gribble, G.W. Recently Discovered Naturally Occurring Heterocyclic Organohalogen Compounds. Heterocycles 2012, 84, 157–207. [Google Scholar] [CrossRef]
- Gribble, G.W. Umweltgifte vom Gabentisch der Natur. Spektrum Wiss. 2005, 6, 38–45. [Google Scholar]
- Wagner, C.; Omari, M.E.; König, G.M. Biohalogenation: Nature’s Way to Synthesize Halogenated Metabolites. J. Nat. Prod. 2009, 72, 540–553. [Google Scholar] [CrossRef]
- Ehrlich, J.; Bartz, Q.R.; Smith, R.M.; Josyln, D.A.; Burkholder, P.R. Chloromycetin, a New Antibiotic from a Soil Actinomycete. Science 1947, 106, 417. [Google Scholar] [CrossRef]
- Duggar, B.M. Aureomycin: A product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 1948, 51, 177–181. [Google Scholar] [CrossRef]
- Kanbe, K.; Okamura, M.; Hattori, S.; Naganawa, H.; Hamada, M.; Okami, Y.; Takeuchi, T. Thienodolin, a New Plant Growth-regulating Substance Produced by a Streptomycete Strain: I. Taxonomy and Fermentation of the Producing Strain, and the Isolation and Characterization of Thienodolin. Biosci. Biotechnol. Biochem. 1993, 57, 632–635. [Google Scholar] [CrossRef] [Green Version]
- Seibold, C.; Schnerr, H.; Rumpf, J.; Kunzendorf, A.; Hatscher, C.; Wage, T.; Ernyei, A.J.; Dong, C.; Naismith, J.H.; van Pée, K.-H. A flavin-dependent tryptophan 6-halogenase and its use in modification of pyrrolnitrin biosynthesis. Biocatal. Biotransf. 2006, 24, 401–408. [Google Scholar] [CrossRef]
- Yeh, E.; Garneau, S.; Walsh, C.T. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 3960–3965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patallo, E.P.; Walter, A.; Milbredt, D.; Thomas, M.; Neumann, M.; Caputi, L.; O’Connor, S.E.; Ludwig-Müller, J.; van Pée, K.-H. Strategies to Produce Chlorinated Indole-3-Acetic Acid and Indole-3-Acetic Acid Intermediates. ChemistrySelect 2017, 2, 11148–11153. [Google Scholar] [CrossRef]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Davidson, S.E.; Davies, N.W.; Smith, J.A.; Dalmais, M.; Bendahmane, A.I.; Quittenden, L.J.; Sutton, L.; Bala, R.K.; Le Signor, C.; et al. Biosynthesis of the Halogenated Auxin, 4-Chloroindole-3-Acetic Acid. Plant Physiol. 2012, 159, 1055–1063. [Google Scholar] [CrossRef] [Green Version]
- McCoy, E.; O’Connor, S.E. Directed Biosynthesis of Alkaloid Analogs in the Medicinal Plant Catharanthus roseus. J. Am. Chem. Soc. 2006, 128, 14276–14277. [Google Scholar] [CrossRef]
- Bartel, B. Auxin Biosynthesis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 51–66. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suzaa, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E. The Tryptophan Conjugates of Jasmonic and Indole-3-Acetic Acids Are Endogenous Auxin Inhibitors. Plant Physiol. 2009, 150, 1310–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grsic, S.; Sauerteig, S.; Neuhaus, K.; Albrecht, M.; Rossiter, J.; Ludwig-Müller, J. Physiological analysis of transgenic Arabidopsis thaliana plants expressing one nitrilase isoform in sense or antisense direction. J. Plant Physiol. 1998, 153, 446–456. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth Stage-Based Phenotypic Analysis of Arabidopsis: A Model for High Throughput Functional Genomics in Plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [Green Version]
- Barazani, O.; Friedman, J. Is IAA the Major Root Growth Factor Secreted from Plant-Growth-Mediating Bacteria? J. Chem. Ecol. 1999, 25, 2397–2406. [Google Scholar] [CrossRef]
- Sarwar, M.; Arshad, M.; Martens, D.; Frankenberger, W.T. Tryptophan-dependent biosynthesis of auxins in soil. Plant Soil 1992, 147, 207–215. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Mizutani, J.; Hasegawa, K.J. Allelopathy of oats. II. Allelochemical effect of L-Tryptophan and its concentration in oat root exudates. J. Chem. Ecol. 1994, 20, 315–319. [Google Scholar] [CrossRef]
- Christian, M.; Hannah, W.B.; Lüthen, H.; Jones, A.M. Identification of auxins by a chemical genomics approach. J. Exp. Bot. 2008, 59, 2757–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaldorf, M.; Ludwig-Müller, J. AM fungi might affect the root morphology of maize by increasing indole-3-butyric acid biosynthesis. Physiol. Plant. 2000, 109, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Davies, P.J. Plant Hormones, 3rd ed.; Kluwer: Dordrecht, The Netherlands, 2004; pp. 1–15. [Google Scholar]
- Reinecke, D.M.; Ozga, J.A.; Magnus, V. Effect of halogen substitution of indole-3-acetic acid on biological activity in pea fruit. Phytochemistry 1995, 40, 1361–1366. [Google Scholar] [CrossRef]
- Dahlke, R.; Fraas, S.; Ullrich, K.K.; Heinemann, K.; Romeiks, M.; Rickmeyer, T.; Klebe, G.; Palme, K.; Lüthen, H.; Steffens, B. Protoplast swelling and hypocotyl growth depend on different auxin signaling pathways. Plant Physiol. 2017, 175, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Lüthen, H. New methods to analyse auxin induced growth II: The swelling reaction of protoplasts—A model system for the analysis of auxin signal transduction? Plant Growth Regul. 2000, 32, 115–122. [Google Scholar] [CrossRef]
- Jayasinghege, C.P.A.; Ozga, J.A.; Nadeau, C.D.; Kaur, H.; Reinecke, D.M. TIR1 auxin receptors are implicated in the differential response to 4-Cl-IAA and IAA in developing pea fruit. J. Exp. Bot. 2019, 70, 1239–1253. [Google Scholar] [CrossRef]
- Runguphan, W.; Qu, X.; O’Connor, S.E. Integrating carbon-halogen bond formation into medicinal plant metabolism. Plant Physiol. 2010, 468, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Glenn, W.S.; Nims, E.; O’Connor, S.E. Reengineering a Tryptophan Halogenase to Preferentially Chlorinate a Direct Alkaloid Precursor. J. Am. Chem. Soc. 2011, 133, 19346–19349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin Biosynthesis: A Simple Two-Step Pathway Converts Tryptophan to Indole-3-Acetic Acid in Plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [Green Version]
- Östin, A.; Kowalyczk, M.; Bhalerao, R.P.; Sandberg, G. Metabolism of Indole-3- Acetic Acid in Arabidopsis. Plant Physiol. 1998, 118, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Böttcher, C.; Dennis, E.G.; Booker, G.W.; Polyak, S.W.; Boss, P.K.; Davies, K. A Novel Tool for Studying Auxin-Metabolism: The Inhibition of Grapevine Indole-3-Acetic Acid-Amido Synthetases by a Reaction Intermediate Analogue. PLoS ONE 2012, 7, e37632. [Google Scholar] [CrossRef] [Green Version]
- Oetiker, J.; Aeschbacher, G. Temperature-sensitive plant cells with shunted indole-3-acetic acid conjugation. Plant Physiol. 1997, 114, 1385–1395. [Google Scholar] [CrossRef] [Green Version]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Wang, R.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouwendal, G.J.A.; Mendes, O.; Wolbert, E.J.H.; de Boer, D.A. Enhanced expression in tobacco of the gene encoding green fluorescent protein by modification of its codon usage. Plant Mol. Biol. 1997, 33, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J. Approaching Cellular and Molecular Resolution of Auxin Biosynthesis and Metabolism. Cold Spring Harb. Perspect. Biol. 2010, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef] [Green Version]
- Bender, J.; Celenza, J.L. Indolic glucosinolates at the crossroads of tryptophan metabolism. Phytochem. Rev. 2008, 8, 25–37. [Google Scholar] [CrossRef]
- Normanly, J. Auxin metabolism. Physiol. Plant. 1997, 100, 431–442. [Google Scholar] [CrossRef]
- Lehmann, T.; Janowitz, T.; Sánchez-Parra, B.; Alonso, M.-M.P.; Trompetter, I.; Piotrowski, M.; Pollmann, S. Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front. Plant Sci. 2017, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Skirycz, A.; Reichelt, M.; Burow, M.; Birkemeyer, C.; Rolcik, J.; Kopka, J.; Zanor, M.I.; Gershenzon, J.; Strnad, M.; Szopa, J.; et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006, 47, 10–24. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Novák, O.; Hényková, E.; Sairanen, I.; Kowalczyk, M.; Pospíšil, T.; Ljung, K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Jülke, S.; Bierfreund, N.M.; Decker, E.L.; Reski, R. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 2009, 181, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Ehmann, A. The van Urk-Salkowski reagent—A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. 1977, 132, 267–276. [Google Scholar] [CrossRef]

| Metabolite | Control | Trp | 4-Cl-Trp | 5-Cl-Trp | 6-Cl-Trp | 7-Cl-Trp |
|---|---|---|---|---|---|---|
| Cl-Trp | − | − | 1.5–4.0 | 2.0–2.5 | 1.3–1.8 | 1.7–2.0 |
| Cl-IAN | − | − | 1.0–6.0 | 1.0–2.0 | 0.3–0.6 | 0.4–0.7 |
| Cl-IAA | − | − | − | 0.25–0.4 | 0.07–0.3 | 0.7–0.9 |
| Cl-IAA-Asp | − | − | + | ++ | + | ++ |
| Line | Cl-Trp | Cl-IAN | Cl-IAA |
|---|---|---|---|
| AtpyrH | 1.5–5.5 | ~0.2 | 0.07–0.09 |
| AtpyrHopt1 | 0.2–2.0 | ~0.1 | 0.07–0.08 |
| AtthdH | ~1.1 | 0.04–0.13 | + |
| AtthdHopt1 | + | ~0.01 | − |
| AtprnA | + | + | − |
| AtprnAopt1 | + | + | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walter, A.; Caputi, L.; O’Connor, S.; van Pée, K.-H.; Ludwig-Müller, J. Chlorinated Auxins—How Does Arabidopsis Thaliana Deal with Them? Int. J. Mol. Sci. 2020, 21, 2567. https://doi.org/10.3390/ijms21072567
Walter A, Caputi L, O’Connor S, van Pée K-H, Ludwig-Müller J. Chlorinated Auxins—How Does Arabidopsis Thaliana Deal with Them? International Journal of Molecular Sciences. 2020; 21(7):2567. https://doi.org/10.3390/ijms21072567
Chicago/Turabian StyleWalter, Antje, Lorenzo Caputi, Sarah O’Connor, Karl-Heinz van Pée, and Jutta Ludwig-Müller. 2020. "Chlorinated Auxins—How Does Arabidopsis Thaliana Deal with Them?" International Journal of Molecular Sciences 21, no. 7: 2567. https://doi.org/10.3390/ijms21072567





