The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle
Abstract
:1. Introduction
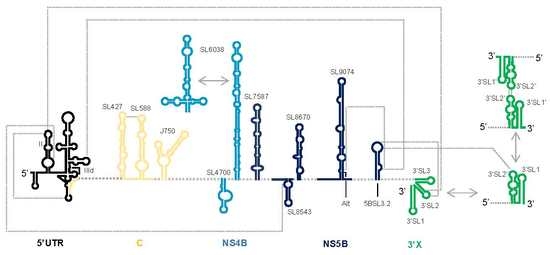
2. The HCV RNA Genome Is a Compact and Resourceful Entity
3. The hepacivirus Life Cycle
4. Translation in HCV Requires Functional Genomic Domains
4.1. Functional RNA Domains Required for Binary Complex Formation
4.2. Structural Basis for the Recognition of eIFs by the HCV IRES
4.3. Dynamic Conformational Tuning of the Translationally Active 80S Complex Mediated by IRES Domains
4.4. Translational Enhancement Mediated by cis-acting RNA Elements and Long Range RNA-RNA Contacts
5. From Translation to Replication. The Role of HCV RNA Circularization
6. Viral RNA Synthesis Is Controlled by High-Order Structures of the Genomic RNA
7. RNA Elements also Control Viral RNA Packaging
8. Genome Circularization Strategies in RNA Viruses Mediated by RNA-RNA Contacts
9. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Kwok, C.K. Dawn of the in vivo RNA structurome and interactome. Biochem. Soc. Trans. 2016, 44, 1395–1410. [Google Scholar] [CrossRef]
- Romero-López, C.; Berzal-Herranz, A. Unmasking the information encoded as structural motifs of viral RNA genomes: A potential antiviral target. Rev. Med. Virol. 2013, 23, 340–354. [Google Scholar] [CrossRef] [Green Version]
- Romero-López, C.; Berzal-Herranz, A. The 5BSL3.2 Functional RNA domain connects distant regions in the hepatitis C virus genome. Front. Microbiol. 2017, 8, 2093. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Siegfried, N.A.; Weeks, K.M. The genetic code as expressed through relationships between mRNA structure and protein function. FEBS Lett. 2013, 587, 1180–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, P. RNA based evolutionary optimization. Orig. Life Evol. Biosph. 1993, 23, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Choo, Q.L.; Kuo, G.; Weiner, A.J.; Overby, L.R.; Bradley, D.W.; Houghton, M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 1989, 244, 359–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alazard-Dany, N.; Denolly, S.; Boson, B.; Cosset, F.L. Overview of HCV life cycle with a special focus on current and possible future antiviral targets. Viruses 2019, 11, 30. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, S.; Bukh, J. Current status and future development of infectious cell-culture models for the major genotypes of hepatitis C virus: Essential tools in testing of antivirals and emerging vaccine strategies. Antivir. Res. 2018, 158, 264–287. [Google Scholar] [CrossRef]
- Taylor, D.R. Evolution of cell culture systems for HCV. Antivir. Ther. 2013, 18, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Sanlés, A.; Ríos-Marco, P.; Romero-López, C.; Berzal-Herranz, A. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front. Microbiol. 2017, 8, 546. [Google Scholar] [CrossRef]
- Ooms, M.; Abbink, T.E.; Pham, C.; Berkhout, B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007, 35, 5253–5261. [Google Scholar] [CrossRef]
- Borgia, S.M.; Hedskog, C.; Parhy, B.; Hyland, R.H.; Stamm, L.M.; Brainard, D.M.; Subramanian, M.G. Identification of a novel hepatitis C virus genotype from Punjab, India: Expanding classification of hepatitis C virus into 8 genotypes. J. Infect. Dis. 2018, 218, 1722–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014, 59, 318–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, N.; Hijikata, M.; Ootsuyama, Y.; Nakagawa, M.; Ohkoshi, S.; Sugimura, T.; Shimotohno, K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 1990, 87, 9524–9528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamizawa, A.; Mori, C.; Fuke, I.; Manabe, S.; Murakami, S.; Fujita, J.; Onishi, E.; Andoh, T.; Yoshida, I.; Okayama, H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 1991, 65, 1105–1113. [Google Scholar] [CrossRef] [Green Version]
- Walewski, J.L.; Keller, T.R.; Stump, D.D.; Branch, A.D. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 2001, 7, 710–721. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Choi, J.; Yen, T.S.; Lu, W.; Strohecker, A.; Govindarajan, S.; Chien, D.; Selby, M.J.; Ou, J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001, 20, 3840–3848. [Google Scholar] [CrossRef] [Green Version]
- Varaklioti, A.; Vassilaki, N.; Georgopoulou, U.; Mavromara, P. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 2002, 277, 17713–17721. [Google Scholar] [CrossRef] [Green Version]
- Boulant, S.; Becchi, M.; Penin, F.; Lavergne, J.P. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 2003, 278, 45785–45792. [Google Scholar] [CrossRef] [Green Version]
- Mauger, D.M.; Golden, M.; Yamane, D.; Williford, S.; Lemon, S.M.; Martin, D.P.; Weeks, K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 3692–3697. [Google Scholar] [CrossRef] [Green Version]
- Fricke, M.; Dunnes, N.; Zayas, M.; Bartenschlager, R.; Niepmann, M.; Marz, M. Conserved RNA secondary structures and long-range interactions in hepatitis C viruses. RNA 2015, 21, 1219–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirakitikulr, N.; Kohlway, A.; Lindenbach, B.D.; Pyle, A.M. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol. Cell 2016, 62, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Ren, S.; Hu, S.; Wang, W.G.; Subramanian, A.; Contreras, D.; Kanagavel, V.; Chung, E.; Ko, J.; Amirtham Jacob Appadorai, R.S.; et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis C virus genome. J. Virol. 2013, 87, 5678–5696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphreys, I.; Fleming, V.; Fabris, P.; Parker, J.; Schulenberg, B.; Brown, A.; Demetriou, C.; Gaudieri, S.; Pfafferott, K.; Lucas, M.; et al. Full-length characterization of hepatitis C virus subtype 3a reveals novel hypervariable regions under positive selection during acute infection. J. Virol. 2009, 83, 11456–11466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patino-Galindo, J.A.; Gonzalez-Candelas, F. Comparative analysis of variation and selection in the HCV genome. Infect. Genet. Evol. 2017, 49, 104–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fricke, M.; Marz, M. Prediction of conserved long-range RNA-RNA interactions in full viral genomes. Bioinformatics 2016, 32, 2928–2935. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Lemon, S.M. Innate immune responses in hepatitis C virus infection. Semin. Immunopathol. 2013, 35, 53–72. [Google Scholar] [CrossRef] [Green Version]
- Han, J.Q.; Barton, D.J. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 2002, 8, 512–525. [Google Scholar] [CrossRef]
- Lohmann, V.; Roos, A.; Korner, F.; Koch, J.O.; Bartenschlager, R. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J. Viral. Hepat. 2000, 7, 167–174. [Google Scholar] [CrossRef]
- Sheridan, I.; Pybus, O.G.; Holmes, E.C.; Klenerman, P. High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression. J. Virol. 2004, 78, 3447–3454. [Google Scholar] [CrossRef] [Green Version]
- Cuevas, J.M.; Torres-Puente, M.; Jimenez-Hernandez, N.; Bracho, M.A.; Garcia-Robles, I.; Wrobel, B.; Carnicer, F.; del Olmo, J.; Ortega, E.; Moya, A.; et al. Genetic variability of hepatitis C virus before and after combined therapy of interferon plus ribavirin. PLoS ONE 2008, 3, e3058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuevas, J.M.; Gonzalez-Candelas, F.; Moya, A.; Sanjuan, R. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J. Virol. 2009, 83, 5760–5764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geller, R.; Estada, U.; Peris, J.B.; Andreu, I.; Bou, J.V.; Garijo, R.; Cuevas, J.M.; Sabariegos, R.; Mas, A.; Sanjuan, R. Highly heterogeneous mutation rates in the hepatitis C virus genome. Nat. Microbiol. 2016, 1, 16045. [Google Scholar] [CrossRef]
- Lavie, M.; Dubuisson, J. Interplay between hepatitis C virus and lipid metabolism during virus entry and assembly. Biochimie 2017, 141, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Xie, Z.; Miao, J.; Ran, J.; Feng, Y.; Xia, X. Regulated entry of hepatitis C virus into hepatocytes. Viruses 2017, 9, 100. [Google Scholar] [CrossRef] [Green Version]
- Grakoui, A.; McCourt, D.W.; Wychowski, C.; Feinstone, S.M.; Rice, C.M. Characterization of the hepatitis C virus-encoded serine proteinase: Determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 1993, 67, 2832–2843. [Google Scholar] [CrossRef] [Green Version]
- Romero-Brey, I.; Merz, A.; Chiramel, A.; Lee, J.Y.; Chlanda, P.; Haselman, U.; Santarella-Mellwig, R.; Habermann, A.; Hoppe, S.; Kallis, S.; et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012, 8, e1003056. [Google Scholar]
- Quinkert, D.; Bartenschlager, R.; Lohmann, V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005, 79, 13594–13605. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Sarnow, P.; Siddiqui, A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993, 67, 3338–3344. [Google Scholar] [CrossRef] [Green Version]
- Ji, H.; Fraser, C.S.; Yu, Y.; Leary, J.; Doudna, J.A. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. USA 2004, 101, 16990–16995. [Google Scholar] [CrossRef] [Green Version]
- Otto, G.A.; Puglisi, J.D. The pathway of HCV IRES-mediated translation initiation. Cell 2004, 119, 369–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pestova, T.V.; Shatsky, I.N.; Fletcher, S.P.; Jackson, R.J.; Hellen, C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998, 12, 67–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lytle, J.R.; Wu, L.; Robertson, H.D. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA 2002, 8, 1045–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sizova, D.V.; Kolupaeva, V.G.; Pestova, T.V.; Shatsky, I.N.; Hellen, C.U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998, 72, 4775–4782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, G.A.; Lukavsky, P.J.; Lancaster, A.M.; Sarnow, P.; Puglisi, J.D. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA 2002, 8, 913–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, K.E.; Waghray, S.; Mortimer, S.A.; Bai, Y.; Doudna, J.A. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure 2011, 19, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Querol-Audi, J.; Mortimer, S.A.; Arias-Palomo, E.; Doudna, J.A.; Nogales, E.; Cate, J.H. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013, 41, 7512–7521. [Google Scholar] [CrossRef]
- Hashem, Y.; des Georges, A.; Dhote, V.; Langlois, R.; Liao, H.Y.; Grassucci, R.A.; Pestova, T.V.; Hellen, C.U.; Frank, J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 2013, 503, 539–543. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Unbehaun, A.; Loerke, J.; Behrmann, E.; Collier, M.; Burger, J.; Mielke, T.; Spahn, C.M. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014, 21, 721–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terenin, I.M.; Dmitriev, S.E.; Andreev, D.E.; Shatsky, I.N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008, 15, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, S.E.; Terenin, I.M.; Andreev, D.E.; Ivanov, P.A.; Dunaevsky, J.E.; Merrick, W.C.; Shatsky, I.N. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 2010, 285, 26779–26787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Park, S.M.; Park, J.H.; Keum, S.J.; Jang, S.K. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011, 30, 2454–2464. [Google Scholar] [CrossRef] [Green Version]
- Jaafar, Z.A.; Oguro, A.; Nakamura, Y.; Kieft, J.S. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, A.M.; Jan, E.; Sarnow, P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 2006, 12, 894–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukiyama-Kohara, K.; Iizuka, N.; Kohara, M.; Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992, 66, 1476–1483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, M.; Ping, L.H.; Rijnbrand, R.C.; Amphlett, E.; Clarke, B.; Rowlands, D.; Lemon, S.M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology 1996, 222, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, K.E.; Waghray, S.; Doudna, J.A. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA 2010, 16, 1559–1569. [Google Scholar] [CrossRef] [Green Version]
- Malygin, A.A.; Kossinova, O.A.; Shatsky, I.N.; Karpova, G.G. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 2013, 41, 8706–8714. [Google Scholar] [CrossRef] [Green Version]
- Perard, J.; Leyrat, C.; Baudin, F.; Drouet, E.; Jamin, M. Structure of the full-length HCV IRES in solution. Nat. Commun. 2013, 4, 1612. [Google Scholar] [CrossRef] [Green Version]
- García-Sacristán, A.; Moreno, M.; Ariza-Mateos, A.; Lopez-Camacho, E.; Jaudenes, R.M.; Vazquez, L.; Gomez, J.; Martin-Gago, J.A.; Briones, C. A magnesium-induced RNA conformational switch at the internal ribosome entry site of hepatitis C virus genome visualized by atomic force microscopy. Nucleic Acids Res. 2015, 43, 565–580. [Google Scholar] [CrossRef] [Green Version]
- Quade, N.; Boehringer, D.; Leibundgut, M.; van den Heuvel, J.; Ban, N. Cryo-EM structure of hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat. Commun. 2015, 6, 7646. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Collier, M.; Loerke, J.; Ismer, J.; Schmidt, A.; Hilal, T.; Sprink, T.; Yamamoto, K.; Mielke, T.; Burger, J.; et al. Molecular architecture of the ribosome-bound hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 2015, 34, 3042–3058. [Google Scholar] [CrossRef] [Green Version]
- Gunther, T. Concentration, compartmentation and metabolic function of intracellular free Mg2+. Magnes Res. 2006, 19, 225–236. [Google Scholar] [PubMed]
- Kieft, J.S.; Zhou, K.; Jubin, R.; Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 2001, 7, 194–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieft, J.S.; Zhou, K.; Jubin, R.; Murray, M.G.; Lau, J.Y.; Doudna, J.A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999, 292, 513–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, D.; Mauro, V.P. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 15385–15389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angulo, J.; Ulryck, N.; Deforges, J.; Chamond, N.; Lopez-Lastra, M.; Masquida, B.; Sargueil, B. Loop IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 2016, 44, 1309–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barria, M.I.; Gonzalez, A.; Vera-Otarola, J.; Leon, U.; Vollrath, V.; Marsac, D.; Monasterio, O.; Perez-Acle, T.; Soza, A.; Lopez-Lastra, M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role on cap-independent translation. Nucleic Acids Res. 2009, 37, 957–971. [Google Scholar] [CrossRef]
- Lukavsky, P.J.; Otto, G.A.; Lancaster, A.M.; Sarnow, P.; Puglisi, J.D. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000, 7, 1105–1110. [Google Scholar]
- Jubin, R.; Vantuno, N.E.; Kieft, J.S.; Murray, M.G.; Doudna, J.A.; Lau, J.Y.; Baroudy, B.M. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 2000, 74, 10430–10437. [Google Scholar] [CrossRef] [Green Version]
- Kolupaeva, V.G.; Pestova, T.V.; Hellen, C.U. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000, 74, 6242–6250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, A.; Chakravarti, A.; Roy, P.; Kar, P.; Siddiqui, O. Frequency of nucleotide sequence variations in the internal ribosome entry site region of hepatitis C virus RNA isolated from responding and non-responding patients with hepatitis C virus genotype 3 infection. Virus Dis. 2016, 27, 251–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berzal-Herranz, A.; Romero-López, C.; Berzal-Herranz, B.; Ramos-Lorente, S. Potential of the other genetic information coded by the viral RNA genomes as antiviral target. Pharmaceuticals 2019, 12, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spahn, C.M.; Kieft, J.S.; Grassucci, R.A.; Penczek, P.A.; Zhou, K.; Doudna, J.A.; Frank, J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 2001, 291, 1959–1962. [Google Scholar] [CrossRef] [Green Version]
- Passmore, L.A.; Schmeing, T.M.; Maag, D.; Applefield, D.J.; Acker, M.G.; Algire, M.A.; Lorsch, J.R.; Ramakrishnan, V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 2007, 26, 41–50. [Google Scholar] [CrossRef]
- Lyons, A.J.; Lytle, J.R.; Gomez, J.; Robertson, H.D. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem-loop II. Nucleic Acids Res. 2001, 29, 2535–2541. [Google Scholar] [CrossRef] [Green Version]
- Lukavsky, P.J.; Kim, I.; Otto, G.A.; Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003, 10, 1033–1038. [Google Scholar] [CrossRef]
- Babaylova, E.; Graifer, D.; Malygin, A.; Stahl, J.; Shatsky, I.; Karpova, G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009, 37, 1141–1151. [Google Scholar] [CrossRef] [Green Version]
- Boehringer, D.; Thermann, R.; Ostareck-Lederer, A.; Lewis, J.D.; Stark, H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure 2005, 13, 1695–1706. [Google Scholar] [CrossRef] [Green Version]
- Lafuente, E.; Ramos, R.; Martínez-Salas, E. Long-range RNA-RNA interactions between distant regions of the hepatitis C virus internal ribosome entry site element. J. Gen. Virol. 2002, 83, 1113–1121. [Google Scholar] [CrossRef]
- Filbin, M.E.; Kieft, J.S. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA 2011, 17, 1258–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pestova, T.V.; Hellen, C.U.; Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell Biol. 1996, 16, 6859–6869. [Google Scholar] [CrossRef] [Green Version]
- Collier, A.J.; Gallego, J.; Klinck, R.; Cole, P.T.; Harris, S.J.; Harrison, G.P.; Aboul-Ela, F.; Varani, G.; Walker, S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat. Struct. Biol. 2002, 9, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Perard, J.; Rasia, R.; Medenbach, J.; Ayala, I.; Boisbouvier, J.; Drouet, E.; Baudin, F. Human initiation factor eIF3 subunit b interacts with HCV IRES RNA through its N-terminal RNA recognition motif. FEBS Lett. 2009, 583, 70–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mercuri, L.; Thomson, E.C.; Hughes, J.; Karayiannis, P. Quasispecies changes with distinctive point mutations in the hepatitis C virus internal ribosome entry site (IRES) derived from PBMCs and plasma. Adv. Virol. 2018, 2018, 4835252. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Barroso-delJesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. The folding of the hepatitis C virus internal ribosome entry site depends on the 3′-end of the viral genome. Nucleic Acids Res. 2012, 40, 11697–11713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Sarnow, P.; Siddiqui, A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 1994, 68, 7301–7307. [Google Scholar] [CrossRef] [Green Version]
- Buratti, E.; Gerotto, M.; Pontisso, P.; Alberti, A.; Tisminetzky, S.G.; Baralle, F.E. In vivo translational efficiency of different hepatitis C virus 5′-UTRs. FEBS Lett. 1997, 411, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Filbin, M.E.; Kieft, J.S. Linking alpha to omega: Diverse and dynamic RNA-based mechanisms to regulate gene expression by 5′-to-3′ communication. F1000Reserach 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Lai, M.M. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 1997, 71, 8698–8706. [Google Scholar] [CrossRef] [Green Version]
- Tsuchihara, K.; Tanaka, T.; Hijikata, M.; Kuge, S.; Toyoda, H.; Nomoto, A.; Yamamoto, N.; Shimotohno, K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 1997, 71, 6720–6726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Lai, M.M. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 1999, 254, 288–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Zhou, K.; Doudna, J.A. Hepatitis C virus 3′UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 2013, 41, 7861–7874. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.; Frederickson, R.M.; Fields, S.; Patel, A.H. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 2001, 75, 1348–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Kato, N.; Cho, M.J.; Shimotohno, K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 1995, 215, 744–749. [Google Scholar] [CrossRef]
- Kolykhalov, A.A.; Feinstone, S.M.; Rice, C.M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 1996, 70, 3363–3371. [Google Scholar] [CrossRef] [Green Version]
- Blight, K.J.; Rice, C.M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 1997, 71, 7345–7352. [Google Scholar] [CrossRef] [Green Version]
- Shetty, S.; Kim, S.; Shimakami, T.; Lemon, S.M.; Mihailescu, M.R. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA 2010, 16, 913–925. [Google Scholar] [CrossRef] [Green Version]
- Cantero-Camacho, A.; Gallego, J. The conserved 3′X terminal domain of hepatitis C virus genomic RNA forms a two-stem structure that promotes viral RNA dimerization. Nucleic Acids Res. 2015, 43, 8529–8539. [Google Scholar] [CrossRef]
- Tuplin, A.; Struthers, M.; Cook, J.; Bentley, K.; Evans, D.J. Inhibition of HCV translation by disrupting the structure and interactions of the viral CRE and 3′ X-tail. Nucleic Acids Res. 2015, 43, 2914–2926. [Google Scholar] [CrossRef]
- Tuplin, A.; Wood, J.; Evans, D.J.; Patel, A.H.; Simmonds, P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 2002, 8, 824–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, S.; Stump, D.D.; Branch, A.D.; Rice, C.M. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 2004, 78, 1352–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friebe, P.; Boudet, J.; Simorre, J.P.; Bartenschlager, R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 2005, 79, 380–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuplin, A.; Struthers, M.; Simmonds, P.; Evans, D.J. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012, 40, 6908–6921. [Google Scholar] [CrossRef]
- Lee, H.; Shin, H.; Wimmer, E.; Paul, A.V. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 2004, 78, 10865–10877. [Google Scholar] [CrossRef] [Green Version]
- Shetty, S.; Stefanovic, S.; Mihailescu, M.R. Hepatitis C virus RNA: Molecular switches mediated by long-range RNA-RNA interactions? Nucleic Acids Res. 2013, 41, 2526–2540. [Google Scholar] [CrossRef] [Green Version]
- Cantero-Camacho, A.; Fan, L.; Wang, Y.X.; Gallego, J. Three-dimensional structure of the 3′X-tail of hepatitis C virus RNA in monomeric and dimeric states. RNA 2017. [Google Scholar] [CrossRef] [Green Version]
- Egger, D.; Wolk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef] [Green Version]
- Paul, D.; Bartenschlager, R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013, 2, 32–48. [Google Scholar] [CrossRef]
- Zhang, J.; Yamada, O.; Sakamoto, T.; Yoshida, H.; Araki, H.; Murata, T.; Shimotohno, K. Inhibition of hepatitis C virus replication by pol III-directed overexpression of RNA decoys corresponding to stem-loop structures in the NS5B coding region. Virology 2005, 342, 276–285. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T. Hepatitis C virus replication. Adv. Exp. Med. Biol. 2017, 997, 199–209. [Google Scholar] [PubMed]
- Tanaka, T.; Sugiyama, K.; Ikeda, M.; Naganuma, A.; Nozaki, A.; Saito, M.; Shimotohno, K.; Kato, N. Hepatitis C virus NS5B RNA replicase specifically binds ribosomes. Microbiol. Immunol. 2000, 44, 543–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Ríos-Marco, P.; Berzal-Herranz, B.; Berzal-Herranz, A. The HCV genome domains 5BSL3.1 and 5BSL3.3 act as managers of translation. Sci. Rep. 2018, 8, 16101. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Berzal-Herranz, A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell Mol. Life Sci. 2012, 69, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Berzal-Herranz, A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 2009, 15, 1740–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-López, C.; Barroso-delJesus, A.; García-Sacristán, A.; Briones, C.; Berzal-Herranz, A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′X-tail region. Nucleic Acids Res. 2014, 42, 567–582. [Google Scholar] [CrossRef] [PubMed]
- Romero-López, C.; Berzal-Herranz, A. Current and emerging themes in the structural analysis of viral RNA genomes: Applications for the development of novel therapeutic drugs. Genom. Comput. Biol. 2015, 1, e15. [Google Scholar] [CrossRef] [Green Version]
- Niepmann, M.; Shalamova, L.A.; Gerresheim, G.K.; Rossbach, O. Signals involved in regulation of hepatitis C virus RNA genome translation and replication. Front. Microbiol. 2018, 9, 395. [Google Scholar] [CrossRef]
- Palau, W.; Masante, C.; Ventura, M.; Di Primo, C. Direct evidence for RNA-RNA interactions at the 3′ end of the hepatitis C virus genome using surface plasmon resonance. RNA 2013, 19, 982–991. [Google Scholar] [CrossRef] [Green Version]
- Rance, E.; Tanner, J.E.; Alfieri, C. Genomic-Scale interaction involving complementary sequences in the hepatitis C virus 5′UTR domain IIa and the RNA-dependent RNA polymerase coding region promotes efficient virus replication. Viruses 2018, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Boerneke, M.A.; Dibrov, S.M.; Gu, J.; Wyles, D.L.; Hermann, T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc. Natl. Acad. Sci. USA 2014, 111, 15952–15957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seth, P.P.; Miyaji, A.; Jefferson, E.A.; Sannes-Lowery, K.A.; Osgood, S.A.; Propp, S.S.; Ranken, R.; Massire, C.; Sampath, R.; Ecker, D.J.; et al. SAR by MS: Discovery of a new class of RNA-binding small molecules for the hepatitis C virus: Internal ribosome entry site IIA subdomain. J. Med. Chem. 2005, 48, 7099–7102. [Google Scholar] [PubMed]
- Friebe, P.; Lohmann, V.; Krieger, N.; Bartenschlager, R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001, 75, 12047–12057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.K.; Kim, C.S.; Lee, S.H.; Jang, S.K. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 2002, 290, 105–112. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.C.; Chang, M.F.; Chang, S.C. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 1999, 73, 7044–7049. [Google Scholar] [CrossRef] [Green Version]
- Diviney, S.; Tuplin, A.; Struthers, M.; Armstrong, V.; Elliott, R.M.; Simmonds, P.; Evans, D.J. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 2008, 82, 9008–9022. [Google Scholar] [CrossRef] [Green Version]
- Kao, C.; Zheng, M.; Rudisser, S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 1999, 5, 1268–1272. [Google Scholar] [CrossRef] [Green Version]
- Ariumi, Y.; Kuroki, M.; Abe, K.; Dansako, H.; Ikeda, M.; Wakita, T.; Kato, N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 2007, 81, 13922–13926. [Google Scholar] [CrossRef] [Green Version]
- Randall, G.; Panis, M.; Cooper, J.D.; Tellinghuisen, T.L.; Sukhodolets, K.E.; Pfeffer, S.; Landthaler, M.; Landgraf, P.; Kan, S.; Lindenbach, B.D.; et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. USA 2007, 104, 12884–12889. [Google Scholar] [CrossRef] [Green Version]
- Oakland, T.E.; Haselton, K.J.; Randall, G. EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J. Virol. 2013, 87, 6625–6634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, D.; Madan, V.; Bartenschlager, R. Hepatitis C virus RNA replication and assembly: Living on the fat of the land. Cell Host Microbe 2014, 16, 569–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ríos-Marco, P.; Romero-López, C.; Berzal-Herranz, A. The cis-acting replication element of the hepatitis C virus genome recruits host factors that influence viral replication and translation. Sci. Rep. 2016, 6, 25729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masante, C.; Jaubert, C.; Palau, W.; Plissonneau, J.; Besnard, L.; Ventura, M.; Di Primo, C. Mutations of the SL2 dimerization sequence of the hepatitis C genome abrogate viral replication. Cell Mol. Life Sci. 2015, 72, 3375–3385. [Google Scholar] [CrossRef]
- Romero-López, C.; Barroso-delJesus, A.; Berzal-Herranz, A. The chaperone-like activity of the hepatitis C virus IRES and CRE elements regulates genome dimerization. Sci. Rep. 2017, 7, 43415. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Rice, C.M. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013, 11, 688–700. [Google Scholar] [CrossRef] [Green Version]
- Shavinskaya, A.; Boulant, S.; Penin, F.; McLauchlan, J.; Bartenschlager, R. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 2007, 282, 37158–37169. [Google Scholar] [CrossRef] [Green Version]
- Shi, G.; Ando, T.; Suzuki, R.; Matsuda, M.; Nakashima, K.; Ito, M.; Omatsu, T.; Oba, M.; Ochiai, H.; Kato, T.; et al. Involvement of the 3′ untranslated region in encapsidation of the hepatitis C virus. PLoS Pathog. 2016, 12, e1005441. [Google Scholar] [CrossRef] [Green Version]
- Nishikawa, F.; Funaji, K.; Fukuda, K.; Nishikawa, S. In vitro selection of RNA aptamers against the HCV NS3 helicase domain. Oligonucleotides 2004, 14, 114–129. [Google Scholar] [CrossRef]
- Dykeman, E.C.; Stockley, P.G.; Twarock, R. Solving a Levinthal’s paradox for virus assembly identifies a unique antiviral strategy. Proc. Natl. Acad. Sci. USA 2014, 111, 5361–5366. [Google Scholar] [CrossRef] [Green Version]
- Stewart, H.; Bingham, R.J.; White, S.J.; Dykeman, E.C.; Zothner, C.; Tuplin, A.K.; Stockley, P.G.; Twarock, R.; Harris, M. Identification of novel RNA secondary structures within the hepatitis C virus genome reveals a cooperative involvement in genome packaging. Sci. Rep. 2016, 6, 22952. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, E.; Brohm, C.; Kallis, S.; Bartenschlager, R.; Pietschmann, T. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 2008, 82, 7034–7046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacini, L.; Graziani, R.; Bartholomew, L.; De Francesco, R.; Paonessa, G. Naturally occurring hepatitis C virus subgenomic deletion mutants replicate efficiently in Huh-7 cells and are trans-packaged in vitro to generate infectious defective particles. J. Virol. 2009, 83, 9079–9093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herold, J.; Andino, R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 2001, 7, 581–591. [Google Scholar] [CrossRef]
- Groft, C.M.; Burley, S.K. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell 2002, 9, 1273–1283. [Google Scholar] [CrossRef]
- Cristofari, G.; Bampi, C.; Wilhelm, M.; Wilhelm, F.X.; Darlix, J.L. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002, 21, 4368–4379. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Lodeiro, M.F.; Luduena, S.J.; Pietrasanta, L.I.; Gamarnik, A.V. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005, 79, 6631–6643. [Google Scholar] [CrossRef] [Green Version]
- Beerens, N.; Kjems, J. Circularization of the HIV-1 genome facilitates strand transfer during reverse transcription. RNA 2010, 16, 1226–1235. [Google Scholar] [CrossRef] [Green Version]
- Khromykh, A.A.; Meka, H.; Guyatt, K.J.; Westaway, E.G. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001, 75, 6719–6728. [Google Scholar] [CrossRef] [Green Version]
- Song, B.H.; Yun, S.I.; Choi, Y.J.; Kim, J.M.; Lee, C.H.; Lee, Y.M. A complex RNA motif defined by three discontinuous 5-nucleotide-long strands is essential for Flavivirus RNA replication. RNA 2008, 14, 1791–1813. [Google Scholar] [CrossRef] [Green Version]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romero-López, C.; Berzal-Herranz, A. The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle. Int. J. Mol. Sci. 2020, 21, 1479. https://doi.org/10.3390/ijms21041479
Romero-López C, Berzal-Herranz A. The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle. International Journal of Molecular Sciences. 2020; 21(4):1479. https://doi.org/10.3390/ijms21041479
Chicago/Turabian StyleRomero-López, Cristina, and Alfredo Berzal-Herranz. 2020. "The Role of the RNA-RNA Interactome in the Hepatitis C Virus Life Cycle" International Journal of Molecular Sciences 21, no. 4: 1479. https://doi.org/10.3390/ijms21041479