Genomic Prediction for Grain Yield and Yield-Related Traits in Chinese Winter Wheat
Abstract
:1. Introduction
2. Results
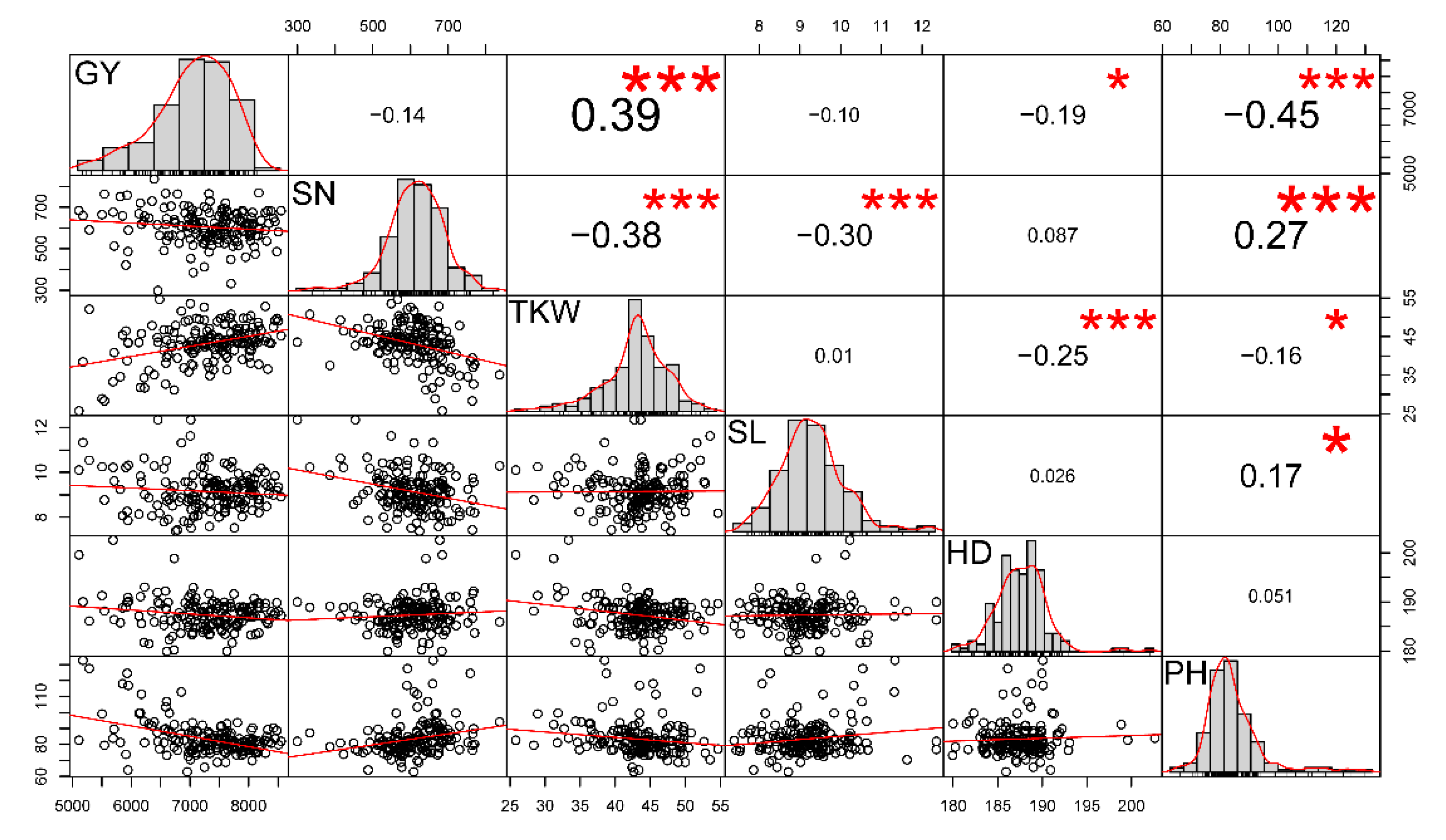
2.1. Phenotypic Evaluation
2.2. Marker Coverage, Genetic Diversity, and Linkage Disequilibrium Analysis
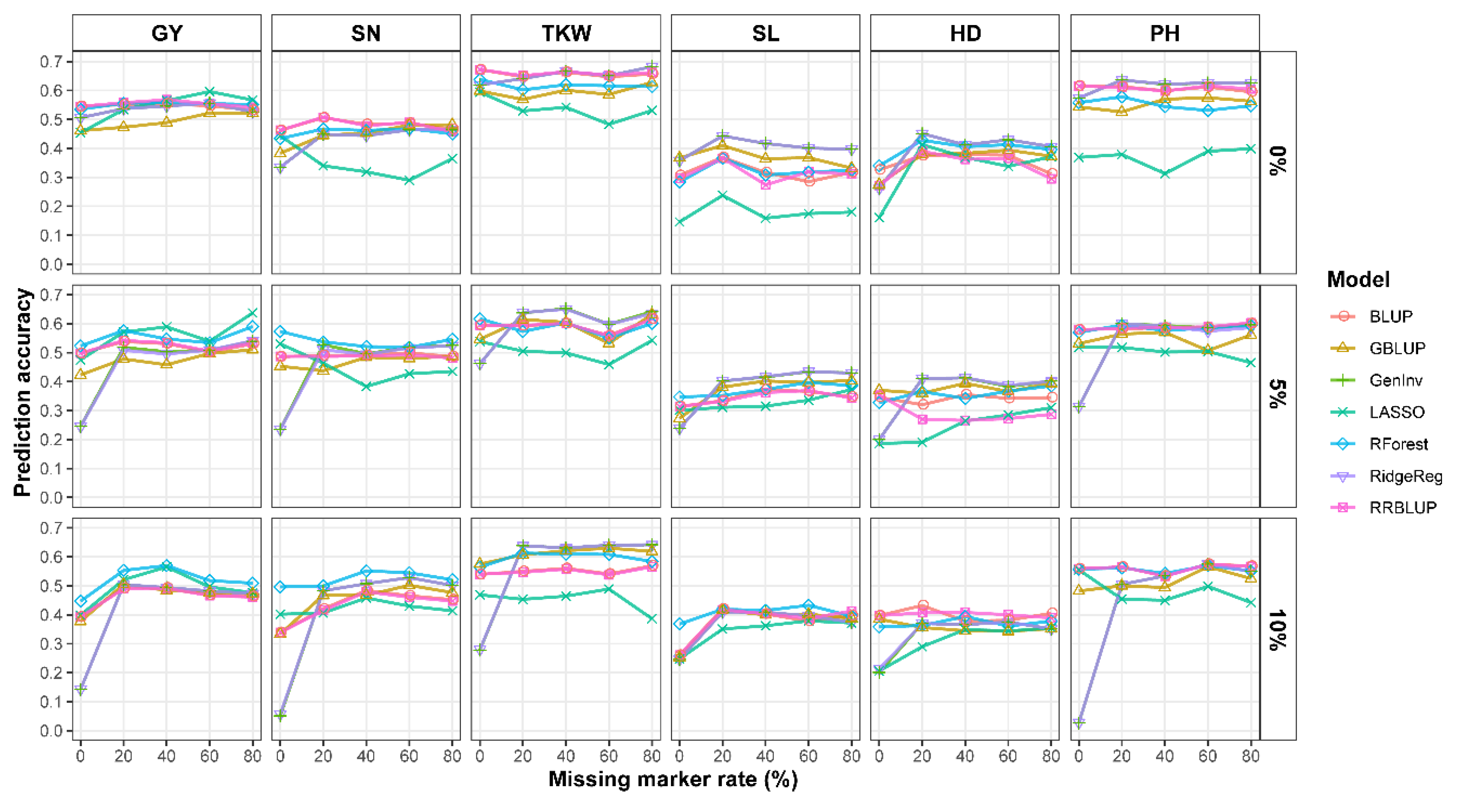
2.3. Prediction Accuracy of Different GS Models under Different Missing Rate and MAF Levels
2.4. Effect of Imputation for Missing Genotypes on GS
2.5. Effect of Significant Markers Detected by GWAS
3. Discussion
3.1. Marker Quality Control, Density, and LD
3.2. Effect of Missing Rate and MAF QC on Prediction Accuracy
3.3. Effect of GS Models on Prediction Accuracy
3.4. Effect of Imputation and GWAS on Prediction Accuracy
4. Materials and Methods
4.1. Plant Materials, Field Trials, and Phenotypic Evaluation
4.2. DNA Extraction, Genotyping, and Quality Control
4.3. Phenotypic Data Analysis and Analysis of Variance (ANOVA)
4.4. Genotypic Data Analysis
4.5. GS Models and Factors Affecting Prediction Accuracy
4.5.1. BLUP Model
4.5.2. GBLUP Model
4.5.3. RRBLUP Model
4.5.4. RidgeReg Model
4.5.5. GenInv Model
4.5.6. LASSO Model
4.5.7. RForest Model
4.6. Imputation for Missing Genotypes
4.7. GWAS-Derived Genomic Selection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| BLUE | Best linear unbiased estimates |
| BLUP | Best linear unbiased predictors |
| GEBV | Genomic estimated breeding values |
| GenInv | Moore-Penrose generalized inverse |
| GBS | Genotyping by sequencing |
| GS | Genomic selection |
| GWAS | Genome-wide association studies |
| GY | Grain yield |
| HD | Heading days |
| LASSO | Least absolute shrinkage and selection operator |
| LD | Linkage disequilibrium |
| MAF | Minor allele frequency |
| PH | Plant height |
| QC | Quality control |
| RForest | Random forest |
| RidgeReg | Ridge regression |
| RRBLUP | Ridge regression best linear unbiased predictors |
| SL | Spike length |
| SN | Spike number |
| SNP | Single nucleotide polymorphism |
| TKW | Thousand-kernel weight |
| TP | Training population |
| VP | Validating population |
References
- FAO FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 2 August 2017).
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating crop genetic gains with genomic selection. Theor. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 2016, 242, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Heslot, N.; Yang, H.P.; Sorrells, M.E.; Jannink, J.L. Genomic selection in plant breeding: A comparison of models. Crop Sci. 2012, 52, 146–160. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Machine Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Rodríguez, P.; Gianola, D.; González-Camacho, J.M.; Crossa, J.; Manès, Y.; Dreisigacker, S. Comparison between linear and non-parametric regression models for genome-enabled prediction in wheat. G3 Genes Genomes Genet. 2012, 2, 1595–1605. [Google Scholar] [CrossRef] [Green Version]
- Charmet, G.; Storlie, E.; Oury, F.X.; Laurent, V.; Beghin, D.; Chevarin, L.; Lapierre, A.; Perretant, M.R.; Rolland, B.; Heumez, E. Genome-wide prediction of three important traits in bread wheat. Mol. Breeding 2014, 34, 1843–1852. [Google Scholar] [CrossRef] [Green Version]
- Jannink, J.L.; Lorenz, A.J.; Iwata, H. Genomic selection in plant breeding: From theory to practice. Brief. Funct. Genomics 2010, 9, 166–177. [Google Scholar] [CrossRef] [Green Version]
- Lozada, D.N.; Mason, R.E.; Sarinelli, J.M.; Brown-Guedira, G. Accuracy of genomic selection for grain yield and agronomic traits in soft red winter wheat. BMC Genet. 2019, 20, 82. [Google Scholar] [CrossRef] [PubMed]
- Norman, A.; Taylor, J.; Edwards, J.; Kuchel, H. Optimising genomic selection in wheat: Effect of marker density, population size and population structure on prediction accuracy. G3 Genes Genomes Genet. 2018, 8, 2889–2899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bresolin, T.; de Magalhães Rosa, G.J.; Valente, B.D.; Espigolan, R.; Gordo, D.G.M.; Braz, C.U.; Fernandes, G.A.; Magalhães, A.F.B.; Garcia, D.A.; Frezarim, G.B. Effect of quality control, density and allele frequency of markers on the accuracy of genomic prediction for complex traits in Nellore cattle. Anim. Prod. Sci. 2019, 59, 48–54. [Google Scholar] [CrossRef] [Green Version]
- Jarquín, D.; Howard, R.; Graef, G.; Lorenz, A. Response surface analysis of genomic prediction accuracy values using quality control covariates in soybean. Evol. Bioinfrom. 2019, 15, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Juliana, P.; Singh, R.P.; Singh, P.K.; Crossa, J.; Huerta-Espino, J.; Lan, C.; Bhavani, S.; Rutkoski, J.E.; Poland, J.A.; Bergstrom, G.C. Genomic and pedigree-based prediction for leaf, stem, and stripe rust resistance in wheat. Theor. Appl. Genet. 2017, 130, 1415–1430. [Google Scholar] [CrossRef] [Green Version]
- Hayes, B.; Panozzo, J.; Walker, C.; Choy, A.; Kant, S.; Wong, D.; Tibbits, J.; Daetwyler, H.; Rochfort, S.; Hayden, M. Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theor. Appl. Genet. 2017, 130, 2505–2519. [Google Scholar] [CrossRef]
- Velu, G.; Crossa, J.; Singh, R.P.; Hao, Y.; Dreisigacker, S.; Perez-Rodriguez, P.; Joshi, A.K.; Chatrath, R.; Gupta, V.; Balasubramaniam, A. Genomic prediction for grain zinc and iron concentrations in spring wheat. Theor. Appl. Genet. 2016, 129, 1595–1605. [Google Scholar] [CrossRef]
- Norman, A.; Taylor, J.; Tanaka, E.; Telfer, P.; Edwards, J.; Martinant, J.P.; Kuchel, H. Increased genomic prediction accuracy in wheat breeding using a large Australian panel. Theor. Appl. Genet. 2017, 130, 2543–2555. [Google Scholar] [CrossRef] [Green Version]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Zhao, D.; Chen, X.; Zhang, Y.; Wang, J. Use of genomic selection and breeding simulation in cross prediction for improvement of yield and quality in wheat (Triticum aestivum L.). Crop J. 2018, 6, 353–365. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, Z.; Liu, G.; Jiang, Y.; Maurer, H.P.; Würschum, T.; Mock, H.P.; Matros, A.; Ebmeyer, E.; Schachschneider, R. Genome-based establishment of a high-yielding heterotic pattern for hybrid wheat breeding. Proc. Natl. Acad. Sci. USA 2015, 112, 15624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; He, Z.; Rasheed, A.; Wen, W.; Yan, J.; Zhang, P.; Wan, Y.; Zhang, Y.; Xie, C.; Xia, X. Genome-wide association mapping of black point reaction in common wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 17, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edriss, V.; Guldbrandtsen, B.; Lund, M.S.; Su, G. Effect of marker-data editing on the accuracy of genomic prediction. J. Anim. Breed. Genet. 2013, 130, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data quality control in genetic case-control association studies. Nat. Protoco. 2010, 5, 1564. [Google Scholar] [CrossRef] [Green Version]
- Elbasyoni, I.S.; Lorenz, A.J.; Guttieri, M.; Frels, K.; Baenziger, P.S.; Poland, J.; Akhunov, E. A comparison between genotyping-by-sequencing and array-based scoring of SNPs for genomic prediction accuracy in winter wheat. Plant Sci. 2018, 270, 123–130. [Google Scholar] [CrossRef]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Sorrells, M. Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Cavanagh, C.R.; Chao, S.; Wang, S.; Huang, B.E.; Stephen, S.; Kiani, S.; Forrest, K.; Saintenac, C.; Brown-Guedira, G.L.; Akhunova, A. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. USA 2013, 110, 8057–8062. [Google Scholar] [CrossRef] [Green Version]
- Allen, A.M.; Winfield, M.O.; Burridge, A.J.; Downie, R.C.; Benbow, H.R.; Barker, G.L.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A. Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 2017, 15, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zhou, H.; Wu, Y.; Li, X.; Zhao, J.; Zuo, T.; Zhang, X.; Zhang, Y.; Liu, S.; Shen, Y. The impact of genetic relationship and linkage disequilibrium on genomic selection. Plos ONE 2015, 10, e0132379. [Google Scholar] [CrossRef] [Green Version]
- Hickey, J.M.; Crossa, J.; Babu, R.; de los Campos, G. Factors affecting the accuracy of genotype imputation in populations from several maize breeding programs. Crop Sci. 2012, 52, 654–663. [Google Scholar] [CrossRef] [Green Version]
- Habier, D.; Tetens, J.; Seefried, F.-R.; Lichtner, P.; Thaller, G. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 2010, 42, 5. [Google Scholar] [CrossRef] [Green Version]
- Roorkiwal, M.; Rathore, A.; Das, R.R.; Singh, M.K.; Jain, A.; Srinivasan, S.; Gaur, P.M.; Chellapilla, B.; Tripathi, S.; Li, Y. Genome-enabled prediction models for yield related traits in chickpea. Front. Plant Sci. 2016, 7, 1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ornella, L.; Singh, S.; Perez, P.; Burgueno, J.; Singh, R.; Tapia, E.; Bhavani, S.; Dreisigacker, S.; Braun, H.J.; Mathews, K.; et al. Genomic prediction of genetic values for resistance to wheat rusts. Plant Gen. 2012, 5, 136–148. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R news 2002, 2, 18–22. [Google Scholar]
- Thavamanikumar, S.; Dolferus, R.; Thumma, B.R. Comparison of genomic selection models to predict flowering time and spike grain number in two hexaploid wheat doubled haploid populations. G3 Genes Genomes Genet. 2015, 5, 1991–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daetwyler, H.D.; Pong-Wong, R.; Villanueva, B.; Woolliams, J.A. The impact of genetic architecture on genome-wide evaluation methods. Genetics 2010, 185, 1021–1031. [Google Scholar] [CrossRef] [Green Version]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin, Germany, 2016; pp. 33–88. [Google Scholar]
- Bernardo, R. Breeding for Quantitative Traits in Plants; Stemma Press: Minneapolis, MN, USA, 2002; pp. 259–299. [Google Scholar]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef] [Green Version]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Statist. Soc. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, P.; Crossa, J.; Rutkoski, J.; Poland, J.; Singh, R.P.; Legarra, A.; Autrique, E.; Campos, G.d.l.; Burgueño, J.; Dreisigacker, S. Single-step genomic and pedigree genotype × environment interaction models for predicting wheat lines in international environments. Plant Genome 2017, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
| Trait 1 | Variance Components (%) | Heritability 3 | ||||
|---|---|---|---|---|---|---|
| Genotype | Environment | G by E Interaction 2 | Random Error | Plot Level | Genotypic Mean Level | |
| GY | 12.12 | 43.00 | 39.39 | 5.50 | 0.42 | 0.85 |
| SN | 34.32 | 24.11 | 36.19 | 5.39 | 0.69 | 0.92 |
| TKW | 41.38 | 27.68 | 23.76 | 7.18 | 0.77 | 0.97 |
| SL | 42.94 | 8.24 | 34.71 | 14.12 | 0.67 | 0.96 |
| HD | 12.64 | 79.29 | 7.26 | 0.81 | 0.81 | 0.97 |
| PH | 60.19 | 11.97 | 23.09 | 4.75 | 0.85 | 0.98 |
| Missing Rate (%) | MAF (%) | ||
|---|---|---|---|
| 0 1 | 5 | 10 | |
| 0 2 | 1442 | 259 | 181 |
| 20 | 8674 | 5343 | 4368 |
| 40 | 9851 | 5513 | 4494 |
| 60 | 10818 | 5635 | 4596 |
| 80 | 11997 | 5725 | 4675 |
| Trait 1 | Scenario | Genomic Selection Model 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| BLUP | GBLUP | GenInv | LASSO | RForest | RidgeReg | RRBLUP | Mean | ||
| GY | QC | 0.531 (0.027) 3 | 0.458 (0.033) | 0.503 (0.029) | 0.5895 (0.022) | 0.547 (0.028) | 0.495 (0.030) | 0.534 (0.027) | 0.522 (0.016) |
| Non-QC 4 | 0.545 (0.030) | 0.461 (0.035) | 0.506 (0.033) | 0.454 (0.029) | 0.535 (0.028) | 0.506 (0.033) | 0.545 (0.030) | 0.507 (0.014) | |
| p-value | 0.3687 | 0.3892 | 0.3926 | 0.0001 | 0.3688 | 0.4662 | 0.3994 | 0.248 | |
| SN | QC | 0.491 (0.026) | 0.484 (0.025) | 0.496 (0.025) | 0.383 (0.030) | 0.521 (0.027) | 0.494 (0.025) | 0.488 (0.026) | 0.480 (0.017) |
| Non-QC | 0.462 (0.030) | 0.383 (0.034) | 0.335 (0.035) | 0.444 (0.033) | 0.434 (0.031) | 0.335 (0.035) | 0.463 (0.031) | 0.408 (0.021) | |
| p-value | 0.2447 | 0.0096 | 0.0002 | 0.0816 | 0.018 | 0.0002 | 0.2671 | 0.011 | |
| TKW | QC | 0.600 (0.028) | 0.605 (0.038) | 0.652 (0.027) | 0.499 (0.036) | 0.603 (0.028) | 0.650 (0.028) | 0.601 (0.028) | 0.601 (0.019) |
| Non-QC | 0.672 (0.025) | 0.598 (0.032) | 0.619 (0.026) | 0.594 (0.027) | 0.638 (0.027) | 0.619 (0.026) | 0.672 (0.025) | 0.630 (0.012) | |
| p-value | 0.0282 | 0.4463 | 0.19 | 0.0189 | 0.1809 | 0.2122 | 0.0298 | 0.116 | |
| SL | QC | 0.373 (0.046) | 0.402 (0.041) | 0.416 (0.041) | 0.315 (0.041) | 0.373 (0.040) | 0.417 (0.040) | 0.362 (0.050) | 0.380 (0.014) |
| Non-QC | 0.307 (0.047) | 0.367 (0.042) | 0.358 (0.042) | 0.146 (0.047) | 0.284 (0.042) | 0.358 (0.042) | 0.296 (0.048) | 0.302 (0.029) | |
| p-value | 0.1595 | 0.2805 | 0.1629 | 0.0041 | 0.0645 | 0.1575 | 0.1721 | 0.020 | |
| HD | QC | 0.355 (0.038) | 0.394 (0.034) | 0.413 (0.033) | 0.264 (0.033) | 0.342 (0.033) | 0.413 (0.033) | 0.266 (0.040) | 0.350 (0.024) |
| Non-QC | 0.326 (0.036) | 0.276 (0.036) | 0.262 (0.040) | 0.161 (0.047) | 0.340 (0.027) | 0.262 (0.040) | 0.272 (0.033) | 0.271 (0.022) | |
| p-value | 0.2912 | 0.0265 | 0.0065 | 0.0412 | 0.471 | 0.0063 | 0.4991 | 0.017 | |
| PH | QC | 0.585 (0.019) | 0.570 (0.020) | 0.593 (0.019) | 0.502 (0.032) | 0.576 (0.027) | 0.591 (0.020) | 0.586 (0.019) | 0.572 (0.012) |
| Non-QC | 0.616 (0.022) | 0.543 (0.030) | 0.574 (0.023) | 0.369 (0.039) | 0.558 (0.027) | 0.574 (0.040) | 0.615 (0.033) | 0.550 (0.032) | |
| p-value | 0.1438 | 0.2283 | 0.2445 | 0.0057 | 0.3201 | 0.2807 | 0.1372 | 0.269 | |
| Mean | QC | 0.489 (0.043) | 0.486 (0.035) | 0.512 (0.039) | 0.425 (0.051) | 0.494 (0.045) | 0.510 (0.039) | 0.473 (0.054) | |
| Non-QC | 0.488 (0.061) | 0.438 (0.049) | 0.442 (0.059) | 0.461 (0.072) | 0.465 (0.056) | 0.442 (0.059) | 0.477 (0.067) | ||
| Trait 1 | Genomic selection Model 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| BLUP | GBLUP | GenInv | LASSO | RForest | RidgeReg | RRBLUP | Mean | |
| GY | 0.517 (0.031) 3 | 0.491 (0.031) | 0.5314 (0.024) | 0.593 (0.022) | 0.577 (0.024) | 0.531 (0.024) | 0.520 (0.031) | 0.537 (0.13) |
| SN | 0.488 (0.025) | 0.477 (0.036) | 0.520 (0.024) | 0.418 (0.029) | 0.569 (0.026) | 0.518 (0.029) | 0.481 (0.026) | 0.496 (0.018) |
| TKW | 0.600 (0.036) | 0.560 (0.043) | 0.636 (0.024) | 0.586 (0.031) | 0.629 (0.03) | 0.636 (0.031) | 0.602 (0.035) | 0.607 (0.011) |
| SL | 0.370 (0.04) | 0.455 (0.041) | 0.489 (0.024) | 0.370 (0.034) | 0.394 (0.036) | 0.494 (0.036) | 0.392 (0.044) | 0.423 (0.021) |
| HD | 0.305 (0.031) | 0.380 (0.030) | 0.381 (0.024) | 0.377 (0.034) | 0.312 (0.057) | 0.401 (0.029) | 0.256 (0.035) | 0.345 (0.02) |
| PH | 0.549 (0.019) | 0.517 (0.0241) | 0.566 (0.024) | 0.450 (0.035) | 0.570 (0.025) | 0.566 (0.02) | 0.547 (0.02) | 0.538 (0.016) |
| Mean | 0.472 (0.046) | 0.480 (0.025) | 0.521 (0.036) | 0.440 (0.036) | 0.509 (0.051) | 0.524 (0.051) | 0.466 (0.051) | |
| Trait 1 | GWAS QTLs | |
|---|---|---|
| Imputed 2 | Non-imputed | |
| GY | 525 | 514 |
| SN | 537 | 576 |
| TKW | 519 | 553 |
| SL | 520 | 509 |
| HD | 508 | 506 |
| PH | 497 | 522 |
| Total | 3106 | 3080 |
| Trait 1 | Imputation 2 | Genomic Selection Model 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| BLUP | GBLUP | GenInv | LASSO | RForest | RidgeReg | RRBLUP | Mean | ||
| GY | Yes | 0.9014 (0.008) 5 | 0.901 (0.008) | 0.887 (0.008) | 0.721 (0.016) | 0.730 (0.020) | 0.887 (0.008) | 0.901 (0.008) | 0.847 (0.031) |
| No | 0.866 (0.01) | 0.712 (0.038) | 0.788 (0.014) | 0.746 (0.017) | 0.727 (0.017) | 0.788 (0.014) | 0.866 (0.010) | 0.785 (0.024) | |
| SN | Yes | 0.923 (0.007) | 0.859 (0.037) | 0.915 (0.007) | 0.673 (0.025) | 0.741 (0.018) | 0.915 (0.007) | 0.923 (0.005) | 0.850 (0.039) |
| No | 0.889 (0.011) | 0.857 (0.018) | 0.872 (0.010) | 0.767 (0.016) | 0.685 (0.023) | 0.872 (0.010) | 0.889 (0.011) | 0.833 (0.029) | |
| TKW | Yes | 0.942 (0.006) | 0.848 (0.034) | 0.938 (0.005) | 0.737 (0.024) | 0.763 (0.021) | 0.938 (0.005) | 0.942 (0.007) | 0.873 (0.034) |
| No | 0.926 (0.005) | 0.79 (0.036) | 0.881 (0.010) | 0.763 (0.018) | 0.733 (0.021) | 0.880 (0.010) | 0.926 (0.005) | 0.843 (0.03) | |
| SL | Yes | 0.932 (0.005) | 0.861 (0.034) | 0.938 (0.005) | 0.628 (0.026) | 0.674 (0.029) | 0.938 (0.005) | 0.932 (0.005) | 0.843 (0.051) |
| No | 0.881 (0.012) | 0.773 (0.044) | 0.841 (0.016) | 0.669 (0.029) | 0.613 (0.034) | 0.840 (0.016) | 0.881 (0.012) | 0.785 (0.04) | |
| HD | Yes | 0.878 (0.011) | 0.792 (0.030) | 0.846 (0.012) | 0.660 (0.026) | 0.648 (0.024) | 0.846 (0.002) | 0.878 (0.011) | 0.793 (0.037) |
| No | 0.873 (0.010) | 0.818 (0.021) | 0.827 (0.013) | 0.728 (0.018) | 0.653 (0.020) | 0.826 (0.013) | 0.873 (0.010) | 0.800 (0.031) | |
| PH | Yes | 0.901 (0.008) | 0.756 (0.038) | 0.846 (0.013) | 0.621 (0.029) | 0.712 (0.024) | 0.846 (0.013) | 0.901 (0.008) | 0.798 (0.040) |
| No | 0.880 (0.011) | 0.800 (0.023) | 0.840 (0.017) | 0.746 (0.023) | 0.664 (0.020) | 0.810 (0.017) | 0.880 (0.011) | 0.803 (0.029) | |
| Mean | Yes | 0.913 (0.010) | 0.836 (0.021) | 0.895 (0.017) | 0.673 (0.019) | 0.711 (0.018) | 0.895 (0.017) | 0.913 (0.100) | |
| No | 0.886 (0.009) | 0.792 (0.020) | 0.842 (0.014) | 0.737 (0.015) | 0.679 (0.019) | 0.836 (0.019) | 0.886 (0.009) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.; Zhang, Y.; Rasheed, A.; Wang, J.; Zhang, L. Genomic Prediction for Grain Yield and Yield-Related Traits in Chinese Winter Wheat. Int. J. Mol. Sci. 2020, 21, 1342. https://doi.org/10.3390/ijms21041342
Ali M, Zhang Y, Rasheed A, Wang J, Zhang L. Genomic Prediction for Grain Yield and Yield-Related Traits in Chinese Winter Wheat. International Journal of Molecular Sciences. 2020; 21(4):1342. https://doi.org/10.3390/ijms21041342
Chicago/Turabian StyleAli, Mohsin, Yong Zhang, Awais Rasheed, Jiankang Wang, and Luyan Zhang. 2020. "Genomic Prediction for Grain Yield and Yield-Related Traits in Chinese Winter Wheat" International Journal of Molecular Sciences 21, no. 4: 1342. https://doi.org/10.3390/ijms21041342





