Gene Expression Analysis and Metabolite Profiling of Silymarin Biosynthesis during Milk Thistle (Silybum marianum (L.) Gaertn.) Fruit Ripening
Abstract
:1. Introduction
2. Results and Discussion
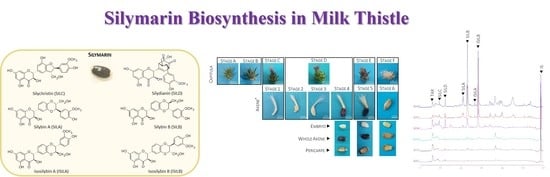
2.1. Morphological Characterization of Milk Thistle Achene Development
2.2. Accumulation Kinetic of SILM Constituents during S. marianum Fruit Development
2.3. Kinetic Study of Selected Enzymatic Activities Related to SILM Biosynthesis
2.4. Expression of Genes Involved in Phenolic Compounds Synthesis
2.4.1. Validation of Reference Genes
2.4.2. Gene Expression Analysis of Candidate Genes
2.5. Relationship between Compounds and Kinetics of ABA Content
3. Materials and Methods
3.1. Plant Materials
3.2. Chemicals
3.3. Phytochemicals Analysis
3.4. Enzymatic Activities
3.4.1. Total Soluble Proteins Extraction and Quantification
3.4.2. PAL Activity
3.4.3. CHS Activity
3.4.4. POX Activity
3.4.5. LAC Activity
3.5. Gene Identification
3.6. Gene Promoter Analysis
3.7. RNA Extraction
3.8. RT-qPCR Analysis
3.9. Validation of Reference Genes
3.10. ABA Extraction and Quantification
3.11. Statistical and Treatment of Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abenavoli, L.; Capasso, R.; Milic, N.; Capasso, F. Milk Thistle in Liver Diseases: Past, Present, Future. Phyther. Res. 2010, 24, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Flora, K.; Martin, H.; Rosen, H.; Benner, K. Clinical reviews Milk Thistle (Silybum marianum) for the Theraply of Liver Disease. Am. J. Gastroenterol. 1998, 93, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Dallio, M.; Loguercio, C. Silymarin/Silybin and chronic liver disease: A marriage of many years. Molecules 2017, 22, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaker, E.; Mahmoud, H.; Mnaa, S. Silymarin, the antioxidant component and Silybum marianum extracts prevent liver damage. Food Chem. Toxicol. 2010, 48, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Agarwal, R. Cosmeceuticals and silibinin. Clin. Dermatol. 2009, 27, 479–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, M.; Ullah, M.A.; Drouet, S.; Younas, M.; Tungmunnithum, D.; Giglioli-Guivarc’h, N.; Hano, C.; Abbasi, B.H. Interactive effects of light and melatonin on biosynthesis of silymarin and anti-inflammatory potential in callus cultures of Silybum marianum (L.) gaertn. Molecules 2019, 24, 1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toklu, H.Z.; Tunali-Akbay, T.; Erkanli, G.; Yüksel, M.; Ercan, F.; Şener, G. Silymarin, the antioxidant component of Silybum marianum, protects against burn-induced oxidative skin injury. Burns 2007, 33, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Trouillas, P.; Marsal, P.; Svobodova, A.; Vosta, J.; Hrba, J.; Lazzaroni, R.; Duroux, J.; Walterova, D. Mechanism of the Antioxidant Action of Silybin and 2,3-Dehydrosilybin Flavonolignans: A Joint Experimental and Theoretical Study. J. Phys. Chem. A 2008, 112, 1054–1063. [Google Scholar] [CrossRef]
- Katiyar, S.K.; Meleth, S.; Sharma, S.D. Silymarin, a flavonoid from milk thistle (Silybum marianum L.), inhibits UV-induced oxidative stress through targeting infiltrating CD11b+cells in mouse skin. Photochem. Photobiol. 2008, 84, 266–271. [Google Scholar] [CrossRef]
- Younas, M.; Drouet, S.; Nadeem, M.; Giglioli-Guivarc’h, N.; Hano, C.; Abbasi, B.H. Differential accumulation of silymarin induced by exposure of Silybum marianum L. callus cultures to several spectres of monochromatic lights. J. Photochem. Photobiol. B Biol. 2018, 184, 61–70. [Google Scholar] [CrossRef]
- Pelter, A.; Hansel, R. The structure of silybin (Silybum substance E6), the first flavonolignan. Tetrahedron Lett. 1968, 1, 2911–2916. [Google Scholar] [CrossRef]
- Nam-Cheol, K.; Graf, T.N.; Sparacino, C.M.; Wani, M.C.; Wall, M.E. Complete isolation and structure identification of hepatoprotective flavonolignans silybins and isosilybins from the medicinal herb milk thistle (Silybum marianum). Org. Biomol. Chem. 2003, 1, 1684–1689. [Google Scholar]
- Kurkin, V.A.; Zapesochnaya, G.G.; Volotsueva, A.V.; Avdeeva, E.V.; Pimenov, K.S. Flavolignans of Silybum marianum fruit. Chem. Nat. Compd. 2001, 37, 315–317. [Google Scholar] [CrossRef]
- Jeong, R.; Lee, D.; Cho, J.; Lee, S.; Kang, H.; Seo, W.; Kang, H.; Kim, J.; Baek, N. Article A New Flavonolignan from the Aerial Parts of Oryza sativa L. Inhibits Nitric oxide Production in RAW 264.7 Macrophage Cells. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 865–870. [Google Scholar] [CrossRef]
- Cardona, M.L.; Garcia, B.; Pedro, R.; Sinisterra, J.F. Flavonoids, Flavonolignans and a phenylpropanoid from Onopordon Corymbosum. Phytochemistry 1990, 29, 629–631. [Google Scholar] [CrossRef]
- Nakajima, Y.; Yun, Y.S.; Kunugi, A. Six new flavonolignans from Sasa veitchii (Carr.) Rehder. Tetrahedron 2003, 59, 8011–8015. [Google Scholar] [CrossRef]
- Bai, N.; He, K.; Roller, M.; Lai, C.S.; Bai, L.; Pan, M.H. Flavonolignans and other constituents from Lepidium meyenii with activities in anti-inflammation and human cancer cell lines. J. Agric. Food Chem. 2015, 63, 2458–2463. [Google Scholar] [CrossRef]
- Wang, J.F.; Yin, G.F.; Zhou, X.J.; Su, J.; Li, Y.; Zhong, H.M.; Duan, G.; Cheng, Y.X. Anti-inflammatory flavonolignans from Hydnocarpus anthelminthica seeds. J. Asian Nat. Prod. Res. 2011, 13, 80–83. [Google Scholar] [CrossRef]
- Karkanis, A.; Bilalis, D.; Efthimiadou, A. Cultivation of milk thistle (Silybum marianum L. Gaertn.), a medicinal weed. Ind. Crops Prod. 2011, 34, 825–830. [Google Scholar] [CrossRef]
- Lv, Y.; Gao, S.; Xu, S.; Du, G.; Zhou, J.; Chen, J.; Road, L.; Chen, J. Spatial organization of silybin biosynthesis in milk thistle (Silybum marianum (L.) Gaertn.). Plant J. 2017, 92, 995–1004. [Google Scholar] [CrossRef] [Green Version]
- Poppe, L.; Petersen, M. Variation in the flavonolignan composition of fruits from different Silybum marianum chemotypes and suspension cultures derived therefrom. Phytochemistry 2016, 131, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Drouet, S.; Abbasi, B.H.; Falguières, A.; Ahmad, W.; Ferroud, C.; Doussot, J.; Vanier, J.R.; Lainé, E.; Hano, C. Single Laboratory Validation of a Quantitative Core Shell-Based LC Separation for the Evaluation of Silymarin Variability and Associated Antioxidant Activity of Pakistani Ecotypes of Milk Thistle (Silybum Marianum, L.). Molecules 2018, 23, 904. [Google Scholar] [CrossRef] [Green Version]
- Davin, L.B.; Wang, H.-B.; Crowell, A.L.; Bedgar, D.L.; Martin, D.M.; Sarkanen, S.; Lewis, N.G. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 1997, 275, 362–367. [Google Scholar] [CrossRef]
- Corbin, C.; Drouet, S.; Markulin, L.; Auguin, D.; Lainé, É.; Davin, L.B.; Cort, J.R.; Lewis, N.G.; Hano, C. A genome-wide analysis of the flax (Linum usitatissimum L.) dirigent protein family: From gene identification and evolution to differential regulation. Plant Mol. Biol. 2018, 97, 73–101. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Corchete, P. Gene expression and flavonolignan production in fruits and cell cultures of Silybum marianum. J. Plant. Physiol. 2016, 192, 111–117. [Google Scholar] [CrossRef]
- Martinelli, T.; Andrzejewska, J.; Salis, M.; Sulas, L. Phenological growth stages of Silybum marianum according to the extended BBCH scale. Ann. Appl. Biol. 2014, 166, 53–66. [Google Scholar] [CrossRef]
- Carrier, D.J.; Crowe, T.; Sokhansanj, S.; Wahab, J.; Branka, B. Milk Thistle, Silybum marianum (L.) Gaertn., flower head development and associated marker compound profile. J. Herbs Spices Med. Plants 2003, 10, 65–74. [Google Scholar] [CrossRef]
- Renouard, S.; Corbin, C.; Lopez, T.; Montguillon, J.; Gutierrez, L.; Lamblin, F.; Lainé, E.; Hano, C. Abscisic acid regulates pinoresinol–lariciresinol reductase gene expression and secoisolariciresinol accumulation in developing flax (Linum usitatissimum L.) seeds. Planta 2012, 235, 85–98. [Google Scholar] [CrossRef]
- Gutierrez, L.; Van Wuytswinkel, O.; Castelain, M.; Bellini, C. Combined networks regulating seed maturation. Trends Plant Sci. 2007, 12, 294–300. [Google Scholar] [CrossRef]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 2002, S15–S45. [Google Scholar] [CrossRef] [Green Version]
- Ciocarlan, A.; Dragalin, I.; Aricu, A.; Ciocarlan, N. Chromatographic analysis of Silybum marianum (L.) gaernt. fatty oil. Chem. J. Mold. 2018, 13, 63–68. [Google Scholar]
- Martinelli, T.; Whittaker, A.; Benedettelli, S.; Carboni, A. The study of flavonolignan association patterns in fruits of diverging Silybum marianum (L.) Gaertn. chemotypes provides new insights into the silymarin biosynthetic pathway. Phytochemistry 2017, 144, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Jamali, A.; Grand, E.; Morreel, K.; Marcelo, P.; Gontier, E.; Dauwe, R. Phenylpropanoid profiling reveals a class of hydroxycinnamoyl glucaric acid conjugates in Isatis tinctoria leaves. Phytochemistry 2017, 144, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Buer, C.S.; Muday, G.K.; Djordjevic, M.A. Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 2007, 145, 478–490. [Google Scholar] [CrossRef] [Green Version]
- Petrussa, E.; Braidot, E.; Zancani, M.; Peresson, C.; Bertolini, A.; Patui, S.; Vianello, A. Plant flavonoids—Biosynthesis, transport and involvement in stress responses. Int. J. Mol. Sci. 2013, 14, 14950–14973. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, L.; Conejero, G.; Castelain, M.; Guénin, S.; Verdeil, J.-L.; Thomasset, B.; Van Wuytswinkel, O. Identification of new gene expression regulators specifically expressed during plant seed maturation. J. Exp. Bot. 2006, 57, 1919–1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.H. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 2010, 61, 2735–2744. [Google Scholar] [CrossRef] [Green Version]
- Hano, C.; Martin, I.; Fliniaux, O.; Legrand, B.; Gutierrez, L.; Arroo, R.R.J.; Mesnard, F.; Lamblin, F.; Lainé, E. Pinoresinol-lariciresinol reductase gene expression and secoisolariciresinol diglucoside accumulation in developing flax (Linum usitatissimum) seeds. Planta 2006, 224, 1291–1301. [Google Scholar] [CrossRef]
- Renouard, S.; Corbin, C.; Lopez, T.; Lamblin, F.; Lainé, E.; Hano, C. Isolation of nuclear proteins from flax (Linum usitatissimum L.) seed coats for gene expression regulation studies. BMC Res. Notes 2012, 5. [Google Scholar] [CrossRef] [Green Version]
- Davin, L.B.; Lewis, N.G. Dirigent Proteins and Dirigent Sites Explain the Mystery of Specificity of Radical Precursor Coupling in Lignan and Lignin Biosynthesis. Plant Physiol. 2000, 123, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Corbin, C.; Renouard, S.; Lopez, T.; Lamblin, F.; Lainé, E.; Hano, C. Identification and characterization of cis-acting elements involved in the regulation of ABA-and/or GA-mediated LuPLR1 gene expression and lignan biosynthesis in flax (Linum usitatissimum L.) cell cultures. J. Plant Physiol. 2013, 170, 516–522. [Google Scholar] [CrossRef] [PubMed]
- Drouet, S.; Leclerc, E.A.; Garros, L.; Tungmunnithum, D.; Kabra, A.; Abbasi, B.H.; Lain, É.; Hano, C. A Green Ultrasound-Assisted Extraction Optimization of the Natural Antioxidant and Anti-Aging Flavonolignans from Milk Thistle Silybum marianum (L.) Gaertn. Fruits for Cosmetic Applications. Antioxidants 2019, 8, 304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hano, C.; Addi, M.; Bensaddek, L.; Crônier, D.; Baltora-Rosset, S.; Doussot, J.; Maury, S.; Mesnard, F.; Chabbert, B.; Hawkins, S.; et al. Differential accumulation of monolignol-derived compounds in elicited flax (Linum usitatissimum) cell suspension cultures. Planta 2006, 223, 975–989. [Google Scholar] [CrossRef]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J.; Hu, H.; Almqvist, J.; Gao, X.; Wang, L. Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE 2015, 10, e0119054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beuerle, T.; Pichersky, E. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal. Biochem. 2002, 302, 305–312. [Google Scholar] [CrossRef] [Green Version]
- Morawski, B.; Lin, Z.; Cirino, P.; Joo, H.; Bandara, G.; Arnold, F.H. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Protein Eng. 2000, 13, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Hu, J.-H.; Guo, C.; Liu, C.-Z. Enhanced laccase production by Trametes versicolor using corn steep liquor as both nitrogen source and inducer. Bioresour. Technol. 2014, 166, 602–605. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, C.-N.; Zheng, H.-Q.; Wu, N.-Y.; Chien, C.-H.; Huang, H.-D.; Lee, T.-Y.; Chiang-Hsieh, Y.-F.; Hou, P.-F.; Yang, T.-Y.; Chang, W.-C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]







| Metabolite | WA1 | WA2 | WA3 | WA4 | WA5 | WA6 |
|---|---|---|---|---|---|---|
| SILM (mg/g DW) | 0.14 ± 0.04 e | 0.42 ± 0.03 d | 0.50 ± 0.04 d | 4.58 ± 0.33 c | 24.20 ± 2.12 b | 52.46 ± 2.73 a |
| SILM (mg/achene) | 0.70 ± 0.21 e | 3.23 ± 0.23 d | 4.78 ± 0.36 d | 174.68 ± 12.46 c | 1100.97 ± 96.55 b | 1528.14 ± 79.59 a |
| ABA (ng/g DW) | 1.83 ± 0.38 e | 5.57 ± 0.63 d | 13.63 ± 1.42 c | 29.10 ± 1.91 b | 48.57 ± 1.56 a | 46.37 ± 2.15 a |
| ABA (ng/achene) | 8.86 ± 1.82 f | 42.86 ± 4.88 e | 129.97 ± 13.53 d | 1108.71 ± 72.69 c | 2209.78 ± 70.99 a | 1350.66 ± 62.60 b |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drouet, S.; Tungmunnithum, D.; Lainé, É.; Hano, C. Gene Expression Analysis and Metabolite Profiling of Silymarin Biosynthesis during Milk Thistle (Silybum marianum (L.) Gaertn.) Fruit Ripening. Int. J. Mol. Sci. 2020, 21, 4730. https://doi.org/10.3390/ijms21134730
Drouet S, Tungmunnithum D, Lainé É, Hano C. Gene Expression Analysis and Metabolite Profiling of Silymarin Biosynthesis during Milk Thistle (Silybum marianum (L.) Gaertn.) Fruit Ripening. International Journal of Molecular Sciences. 2020; 21(13):4730. https://doi.org/10.3390/ijms21134730
Chicago/Turabian StyleDrouet, Samantha, Duangjai Tungmunnithum, Éric Lainé, and Christophe Hano. 2020. "Gene Expression Analysis and Metabolite Profiling of Silymarin Biosynthesis during Milk Thistle (Silybum marianum (L.) Gaertn.) Fruit Ripening" International Journal of Molecular Sciences 21, no. 13: 4730. https://doi.org/10.3390/ijms21134730