SpotLight Proteomics—A IgG-Enrichment Phenotype Profiling Approach with Clinical Implications
Abstract
:1. Introduction
2. Results
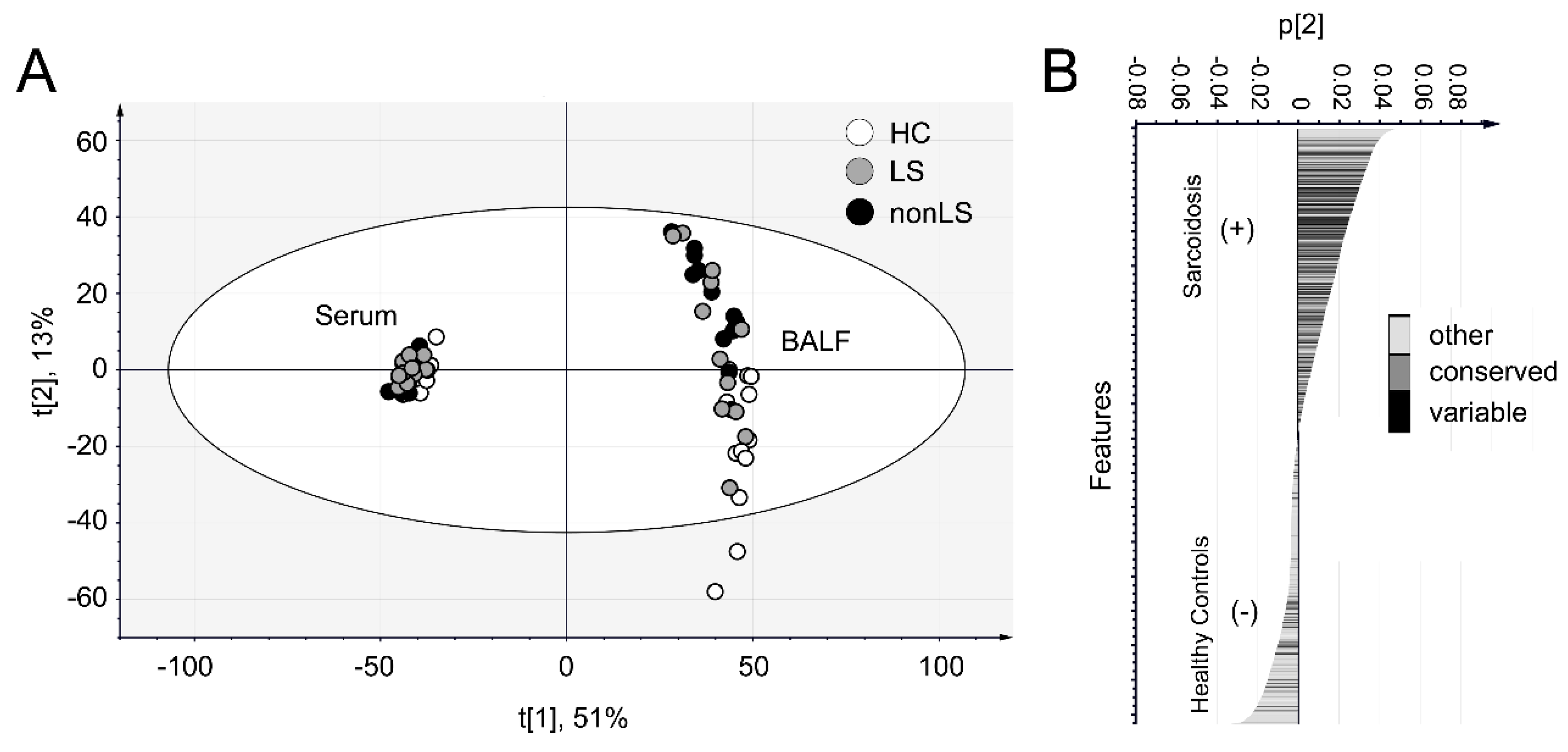
2.1. Data Overview
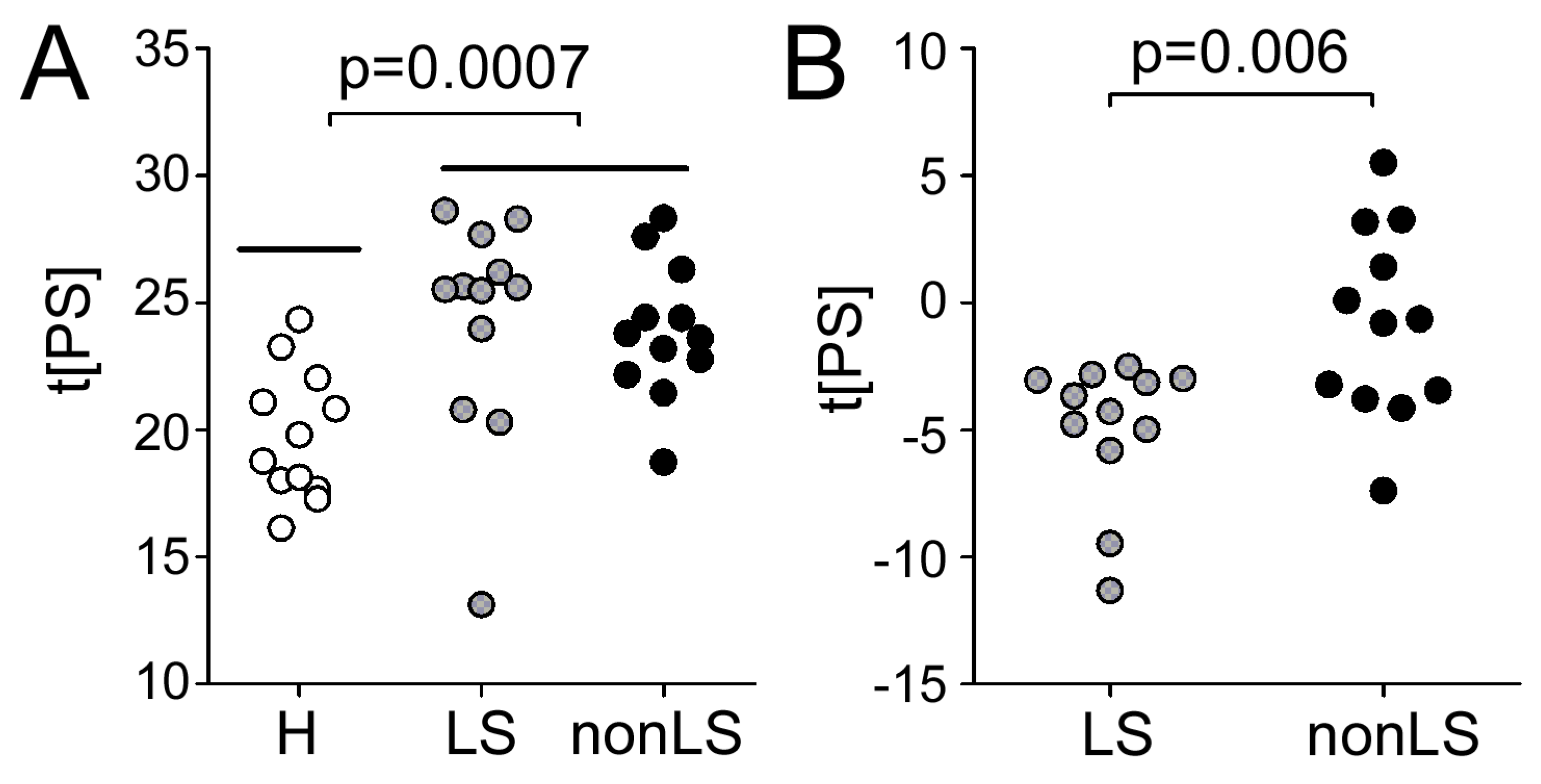
2.2. Differences Between Controls and Sarcoidosis Patients
2.3. Differences Between LS and non-LS Patients
2.4. Predicting Sarcoidosis Disease Status in Serum via Information Obtained from BALF
3. Discussion
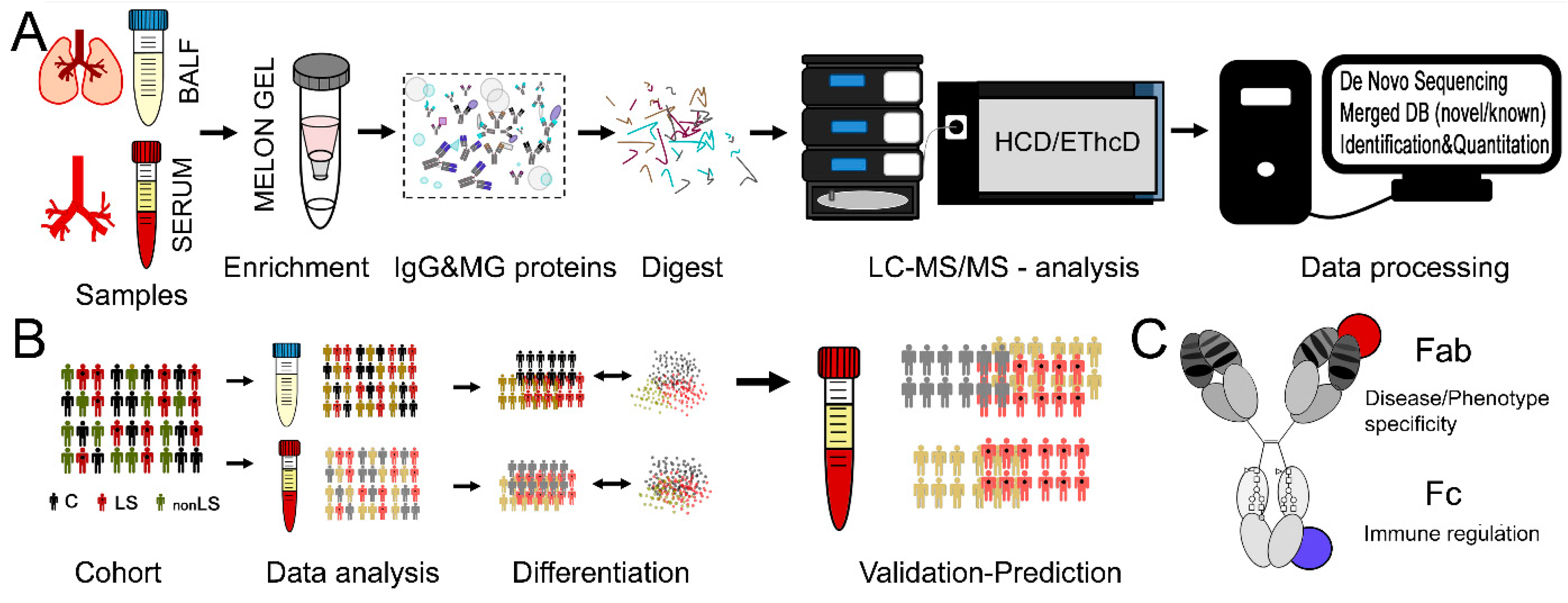
4. Material and Methods
4.1. Subject Information
4.2. Sample Preparations
4.3. LC-MS/MS Analysis
4.4. Protein and Peptide Identification and Quantification
4.5. Fc-Glycopeptide Identification and Quantification
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAMTS10 | A disintegrin and metalloproteinase with thrombospondin motifs 10 |
| ADAMTS19 | A disintegrin and metalloproteinase with thrombospondin motifs 19 |
| ADAMTS6 | A disintegrin and metalloproteinase with thrombospondin motifs 6 |
| AGAP3 | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 3 |
| ALB | Serum albumin |
| ANXA2 | Annexin A2 |
| APC | Adenomatous polyposis coli protein |
| APOA1 | Apolipoprotein A-I |
| APOA2 | Apolipoprotein A-2 |
| APOB | Apolipoprotein B |
| AQR | RNA helicase aquarius |
| ARG1 | Arginase-1 |
| ATIC | Bifunctional purine biosynthesis protein PURH |
| ATP4A | Potassium-transporting ATPase alpha chain 1 |
| C2 | Complement 2 |
| C3 | Complement 3 |
| C8G | Complement component C8 gamma chain |
| CASP14 | Caspase-14 |
| CAT | Catalase |
| CAPN5 | Calpain-5, PLG, Plasminogen |
| CD109 | CD109 antigen |
| CD55 | Complement decay-accelerating factor |
| CFB | Complement factor B |
| CFI | Complement factor I |
| CHIT1 | Chitotriosidase-1 |
| CLSTN1 | Calsyntenin-1 |
| COPB1 | Coatomer subunit beta |
| CST6 | Cystatin-M |
| CSTA | Cystatin-A |
| CSTB | Cystatin-B |
| CTSH | Pro-cathepsin H |
| CTSS | Cathepsin S |
| DCD | Dermcidin |
| DUSP14 | Dual specificity protein phosphatase 14 |
| DYNC2H1 | Cytoplasmic dynein 2 heavy chain 1 |
| ERCC6L | DNA excision repair protein ERCC-6-like |
| F11R | Junctional adhesion molecule A |
| F2 | Prothrombin |
| FABP4 | Fatty acid-binding protein, adipocyte |
| FETUB | Fetuin-B |
| FKBP1A | Peptidyl-prolyl cis-trans isomerase |
| GAA | Lysosomal alpha-glucosidase |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GCHFR | GTP cyclohydrolase 1 feedback regulatory protein |
| GIG25 | Serpin peptidase inhibitor |
| GSTP1 | Glutathione S-transferase P |
| GSN | Gelsolin |
| HBB | Hemoglobin subunit beta |
| HEXDC | Hexosaminidase D |
| HMHA1 | Rho GTPase-activating protein 45 |
| HSPB1 | Heat shock protein beta-1 |
| HPX | Hemopexin |
| INSR | Insulin receptor |
| KIF21B | Kinesin-like protein KIF21B |
| KLKB1 | Plasma kallikrein |
| LTF | Lactotransferrin |
| NPC2 | NPC intracellular cholesterol transporter 2 homolog a |
| MAPK8IP2 | C-Jun-amino-terminal kinase-interacting protein 2 |
| MICAL1 | [F-actin]-monooxygenase MICAL1 |
| MYO3B | Myosin-IIIb |
| MYRF | Myelin regulatory factor |
| NAPSA | Napsin-A |
| PARK7 | Protein/nucleic acid deglycase DJ-1 |
| PEBP1 | Phosphatidylethanolamine-binding protein 1 |
| PIP | Prolactin-inducible protein |
| PLCL1 | Inactive phospholipase C-like protein 1 |
| PTPRQ | Phosphatidylinositol phosphatase, PTPRQ |
| RAB26 | Ras-related protein Rab-26 |
| PREX1 | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein |
| PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 |
| RARRES3 | Retinoic acid receptor responder protein 3 |
| RASEF | Ras and EF-hand domain-containing protein |
| RGS2 | Regulator of G-protein signaling 2 |
| SCGB1A1 | Uteroglobin |
| SERPINB3 | Serpin B3 |
| SERPINB10 | Serpin B10 |
| SERPINB12 | Serpin B12 |
| SLPI | Antileukoproteinase |
| SOD1 | Superoxide dismutase [Cu-Zn] |
| SPINT2 | Kunitz-type protease inhibitor 2 |
| TINAG | Tubulointerstitial nephritis antigen |
| TLL1 | Tolloid-like protein 1 |
| TMEM55A | Type 2 phosphatidylinositol 4,5-bisphosphate 4-phosphatase |
| TMPRSS3 | Transmembrane protease serine 3 |
| TXN | Thioredoxin |
| USP16 | Ubiquitin carboxyl-terminal hydrolase 16 |
| USP43 | Ubiquitin carboxyl-terminal hydrolase 43 |
| WFDC2 | WAP four-disulfide core domain protein 2 |
| WFIKKN2 | WAP, Kazal, immunoglobulin, Kunitz and NTR domain-containing protein 2 |
References
- Iannuzzi, M.C.; Rybicki, B.A.; Teirstein, A.S. Sarcoidosis. N. Engl. J. Med. 2007, 357, 2153–2165. [Google Scholar] [CrossRef]
- Grunewald, J.; Eklund, A. Lofgren’s syndrome: Human leukocyte antigen strongly influences the disease course. Am. J. Respir. Crit. Care Med. 2009, 179, 307–312. [Google Scholar] [CrossRef]
- Fang, C.; Huang, H.; Xu, Z. Immunological evidence for the role of mycobacteria in sarcoidosis: A meta-analysis. PLoS ONE 2016, 11, e0154716. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, G.; Valentini, D.; Rao, M.; Wahlstrom, J.; Grunewald, J.; Larsson, L.O.; Brighenti, S.; Dodoo, E.; Zumla, A.; Maeurer, M. Humoral immune profiling of mycobacterial antigen recognition in sarcoidosis and lofgren’s syndrome using high-content peptide microarrays. Int J. Infect. Dis 2017, 56, 167–175. [Google Scholar] [CrossRef]
- Wahlstrom, J.; Dengjel, J.; Winqvist, O.; Targoff, I.; Persson, B.; Duyar, H.; Rammensee, H.G.; Eklund, A.; Weissert, R.; Grunewald, J. Autoimmune t cell responses to antigenic peptides presented by bronchoalveolar lavage cell hla-dr molecules in sarcoidosis. Clin. Immunol. 2009, 133, 353–363. [Google Scholar] [CrossRef]
- Haggmark, A.; Hamsten, C.; Wiklundh, E.; Lindskog, C.; Mattsson, C.; Andersson, E.; Lundberg, I.E.; Gronlund, H.; Schwenk, J.M.; Eklund, A.; et al. Proteomic profiling reveals autoimmune targets in sarcoidosis. Am. J. Respir. Crit. Care Med. 2015, 191, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, S.L.; Zhang, B.; Rutishauser, D.; Aarsland, D.; Zubarev, R.A. Spotlight proteomics: Uncovering the hidden blood proteome improves diagnostic power of proteomics. Sci. Rep. 2017, 7, 41929. [Google Scholar] [CrossRef]
- Fernandes-Cerqueira, C.; Renard, N.; Notarnicola, A.; Wigren, E.; Graslund, S.; Zubarev, R.A.; Lundberg, I.E.; Lundstrom, S.L. Patients with anti-jo1 antibodies display a characteristic igg fc-glycan profile which is further enhanced in anti-jo1 autoantibodies. Sci. Rep. 2018, 8, 17958. [Google Scholar] [CrossRef]
- Lundstrom, S.L.; Fernandes-Cerqueira, C.; Ytterberg, A.J.; Ossipova, E.; Hensvold, A.H.; Jakobsson, P.J.; Malmstrom, V.; Catrina, A.I.; Klareskog, L.; Lundberg, K.; et al. Igg antibodies to cyclic citrullinated peptides exhibit profiles specific in terms of igg subclasses, fc-glycans and a fab-peptide sequence. PLoS ONE 2014, 9, e113924. [Google Scholar] [CrossRef]
- Heyder, T.; Wiklundh, E.; Eklund, A.; James, A.; Grunewald, J.; Zubarev, R.A.; Lundstrom, S.L. Altered fc galactosylation in igg4 is a potential serum marker for chronic lung disease. Erj. Open Res. 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Salazar, A.; Mana, J.; Pinto, X.; Argimon, J.M.; Hurtado, I.; Pujol, R. Corticosteroid therapy increases hdl-cholesterol concentrations in patients with active sarcoidosis and hypoalphalipoproteinemia. Clin. Chim. Acta 2002, 320, 59–64. [Google Scholar] [CrossRef]
- Bargagli, E.; Rosi, E.; Pistolesi, M.; Lavorini, F.; Voltolini, L.; Rottoli, P. Increased risk of atherosclerosis in patients with sarcoidosis. Pathobiology 2017, 84, 258–263. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Kotur-Stevuljevic, J.; Stefanovic, A.; Jelic-Ivanovic, Z.; Spasic, S.; Videnovic-Ivanov, J.; Vucinic-Mihailovic, V.; Ilic, J. Dyslipidemia and oxidative stress in sarcoidosis patients. Clin. Biochem. 2012, 45, 677–682. [Google Scholar] [CrossRef]
- Chen, E.S.; Song, Z.; Willett, M.H.; Heine, S.; Yung, R.C.; Liu, M.C.; Groshong, S.D.; Zhang, Y.; Tuder, R.M.; Moller, D.R. Serum amyloid a regulates granulomatous inflammation in sarcoidosis through toll-like receptor-2. Am. J. Respir. Crit. Care Med. 2010, 181, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Agostini, C.; Garbisa, S.; Trentin, L.; Zambello, R.; Fastelli, G.; Onisto, M.; Cipriani, A.; Festi, G.; Casara, D.; Semenzato, G. Pulmonary alveolar macrophages from patients with active sarcoidosis express type iv collagenolytic proteinase. An enzymatic mechanism for influx of mononuclear phagocytes at sites of disease activity. J. Clin. Invest. 1989, 84, 605–612. [Google Scholar] [CrossRef]
- Karimi, R.; Tornling, G.; Grunewald, J.; Eklund, A.; Skold, C.M. Cell recovery in bronchoalveolar lavage fluid in smokers is dependent on cumulative smoking history. PLoS ONE 2012, 7, e34232. [Google Scholar] [CrossRef] [PubMed]
- Statement on Sarcoidosis. Joint statement of the american thoracic society (ats), the european respiratory society (ers) and the world association of sarcoidosis and other granulomatous disorders (wasog) adopted by the ats board of directors and by the ers executive committee, february 1999. Am. J. Respir. Crit. Care Med. 1999, 160, 736–755. [Google Scholar]
- Chi, H.; Chen, H.; He, K.; Wu, L.; Yang, B.; Sun, R.X.; Liu, J.; Zeng, W.F.; Song, C.Q.; He, S.M.; et al. Pnovo+: De novo peptide sequencing using complementary hcd and etd tandem mass spectra. J. Proteome Res. 2013, 12, 615–625. [Google Scholar] [CrossRef]
- Zhang, B.; Kall, L.; Zubarev, R.A. Demix-q: Quantification-centered data processing workflow. Mol. Cell Proteom. 2016, 15, 1467–1478. [Google Scholar] [CrossRef]
- Zhang, B.; Pirmoradian, M.; Chernobrovkin, A.; Zubarev, R.A. Demix workflow for efficient identification of cofragmented peptides in high resolution data-dependent tandem mass spectrometry. Mol. Cell Proteom. 2014, 13, 3211–3223. [Google Scholar] [CrossRef]
- Lundstrom, S.L.; Yang, H.; Lyutvinskiy, Y.; Rutishauser, D.; Herukka, S.K.; Soininen, H.; Zubarev, R.A. Blood plasma igg fc glycans are significantly altered in alzheimer’s disease and progressive mild cognitive impairment. J. Alzheimers Dis. 2014, 38, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Royle, L.; Campbell, M.P.; Radcliffe, C.M.; White, D.M.; Harvey, D.J.; Abrahams, J.L.; Kim, Y.G.; Henry, G.W.; Shadick, N.A.; Weinblatt, M.E.; et al. Hplc-based analysis of serum n-glycans on a 96-well plate platform with dedicated database software. Anal. Biochem. 2008, 376, 1–12. [Google Scholar] [CrossRef] [PubMed]



| Type | Correlation | Type | Process/Function | FDR | Gene Name |
|---|---|---|---|---|---|
| Process | Sarcoidosis | Complement | regulation of complement activation | 2.0 × 10−3 | C2, C3, C8G, CFB, CFI, F2 |
| regulation of humoral immune response | 2.0 × 10−3 | C2, C3, C8G, CFB, CFI, F2, HPX | |||
| complement activation | 7.2 × 10−3 | C2, C3, C8G, CFB, CFI | |||
| complement activation, classical pathway | 1.7 × 10−2 | C2, C3, C8G, CFI | |||
| complement activation, alternative pathway | 1.9 × 10−2 | C3, C8G, CFB | |||
| Effector | regulation of immune effector process | 4.0 × 10−2 | APOA1, APOA2, ARG1, C2, C3, C8G, CFB, CFI, F2, HPX | ||
| Neutrophil | neutrophil activation | 2.4 × 10−3 | ARG1, C3, CHIT1, COPB1, GAA, GIG25, GSN, HBB, HMHA1, LTF, NPC2, PREX1, PSMD1, SERPINB10, SERPINB3 | ||
| neutrophil mediated immunity | 2.4 × 10−3 | ARG1, C3, CHIT1, COPB1, F2, GAA, GIG25, GSN, HBB, HMHA1, LTF, NPC2, PSMD1, SERPINB10, SERPINB3 | |||
| Lipid | cholesterol efflux | 7.2 × 10−3 | APOA1, APOA2, APOB, NPC2 | ||
| chylomicron remodeling | 7.6 × 10−3 | APOA1, APOA2, APOB | |||
| chylomicron assembly | 1.2 × 10−2 | APOA1, APOA2, APOB | |||
| plasma lipoprotein particle clearance | 2.6 × 10−2 | APOA1, APOA2, APOB, NPC2 | |||
| negative regulation of very-low-density lipoprotein particle remodeling | 3.8 × 10−2 | APOA1, APOA2 | |||
| Controls | Inhibitors | negative regulation of catalytic activity | 3.7 × 10−8 | ANXA2, APC, CD109, CST6, CSTA, CSTB, F11R, FABP4, FETUB, FKBP1A, GAPDH, GCHFR, GSTP1, HSPB1, MICAL1, PARK7, PEBP1, RGS2, SCGB1A1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |
| negative regulation of hydrolase activity | 5.3 × 10−8 | CD109, CST6, CSTA, CSTB, FETUB, GAPDH, MICAL1, PARK7, PEBP1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |||
| negative regulation of endopeptidase activity | 7.8 × 10−8 | CD109, CST6, CSTA, CSTB, FETUB, GAPDH, MICAL1, PARK7, PEBP1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |||
| negative regulation of proteolysis | 7.6 × 10−7 | CD109, CD55, CST6, CSTA, CSTB, FETUB, GAPDH, MICAL1, PARK7, PEBP1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |||
| Function | Sarcoidosis | Peptidases | endopeptidase activity | 2.1 × 10−3 | ADAMTS10, ADAMTS19, ADAMTS6, C2, C3, CAPN5, CFB, CFI, F2, LTF, PLG, TLL1, TMPRSS3, USP16 |
| peptidase activity, acting on L-amino acid peptides | 5.8 × 10−3 | ADAMTS10, ADAMTS19, ADAMTS6, C2, C3, CAPN5, CFB, CFI, F2, LTF, PLG, TLL1, TMPRSS3, USP16, USP43 | |||
| hydrolase activity | 2.6 × 10−2 | ADAMTS10, ADAMTS19, ADAMTS6, AGAP3, AQR, ARG1, ATIC, ATP4A, C2, C3, CAPN5, CFB, CFI, CHIT1, DUSP14, DYNC2H1, ERCC6L, F2, GAA, HEXDC, KIF21B, LTF, MYO3B, PLCL1, PLG, PTPRQ, RAB26, RARRES3, RASEF, TLL1, TMEM55A, TMPRSS3, USP16, USP43 | |||
| serine-type endopeptidase activity | 4.3 × 10−3 | C2, C3, CFB, CFI, F2, LTF, PLG, TLL1, TMPRSS3 | |||
| Cholesterol | cholesterol transporter activity | 4.3 × 10−3 | APOA1, APOA2, APOB, NPC2 | ||
| cholesterol binding | 2.6 × 10−2 | APOA1, APOA2, APOB, NPC2 | |||
| Lipid | high-density lipoprotein particle receptor binding | 2.6 × 10−2 | APOA1, APOA2 | ||
| apolipoprotein receptor binding | 4.1 × 10−2 | APOA1, APOA2 | |||
| lipoprotein particle receptor binding | 4.1 × 10−2 | APOA1, APOA2, APOB | |||
| phosphatidylcholine-sterol O-acyltransferase activator activity | 4.1 × 10−2 | APOA1, APOA2 | |||
| Amyloid | amyloid-beta binding | 3.7 × 10−2 | APOA1, CLSTN1, INSR, MAPK8IP2 | ||
| Controls | Inhibitors | endopeptidase inhibitor activity | 6.6 × 10−7 | CD109, CST6, CSTA, CSTB, FETUB, GAPDH, PEBP1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |
| enzyme inhibitor activity | 6.6 × 10−7 | ANXA2, CD109, CST6, CSTA, CSTB, FETUB, GAPDH, GCHFR, HSPB1, PEBP1, SCGB1A1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |||
| peptidase regulator activity | 6.6 × 10−7 | CD109, CST6, CSTA, CSTB, CTSH, FETUB, GAPDH, PEBP1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |||
| serine-type endopeptidase inhibitor activity | 2.1 × 10−4 | CD109, PEBP1, SERPINB12, SLPI, SPINT2, WFDC2, WFIKKN2 | |||
| cysteine-type endopeptidase inhibitor activity | 1.8 × 10−3 | CST6, CSTA, CSTB, FETUB, WFDC2 | |||
| aspartic-type endopeptidase inhibitor activity | 1.3 × 10−2 | GAPDH, WFDC2 | |||
| phospholipase A2 inhibitor activity | 1.7 × 10−2 | ANXA2, SCGB1A1 | |||
| protease binding | 3.3 × 10−2 | ANXA2, CST6, CSTA, CSTB, PARK7 | |||
| Antioxidant | antioxidant activity | 8.4 × 10−4 | ALB, CAT, GSTP1, PARK7, SOD1, TXN | ||
| Peptidases | cysteine-type endopeptidase activity | 3.6 × 10−2 | CASP14, CTSH, CTSS, TINAG | ||
| peptidase activity | 3.6 × 10−2 | CASP14, CTSH, CTSS, DCD, KLKB1, MYRF, NAPSA, PARK7, PIP, PLG, TINAG | |||
| endopeptidase activity | 4.7 × 10−2 | CASP14, CTSH, CTSS, KLKB1, NAPSA, PIP, PLG, TINAG |
| H a | LS b | Non-LS c | |
|---|---|---|---|
| Age (year) | 27 ± 3 | 42 ± 7 | 43 ± 9 |
| Smoking (never/ex) | 12/0 | 8/3 | 9/3 |
| Gender (male/female) | 12/0 | 12/0 | 12/0 |
| X-ray stage I/II/III/IVd | NM e | 5/6/0/0 | 3/9/0/0 |
| FVCf% of predictegf | 114 ± 11 | 84 ± 16 | 90 ± 14 |
| FEV(1)hg% of predicted | 107 ± 9 | 86 ± 16 | 86 ± 15 |
| FEV(1)/FVCi | 79 ± 7 | 78 ± 7 | 70 ± 6 |
| Macrophages % * | 86 ± 9 | 72 ± 21 | 69 ± 15 |
| Lymphocytes % * | 9 ± 5 | 26 ± 21 | 28 ± 16 |
| Neutrophils % * | 2 ± 4 | 2 ± 1 | 1 ± 1 |
| Eosinophils % * | 0 | 0 | 2 ± 4 |
| CD4/CD8 | NM | 9 ± 7 | 6 ± 6 |
| Va2.3j% | NM | 18 ± 15 | 8 ± 8 |
| CRPkmg/L | NM | 16 ± 26 | 3 ± 2 |
| ACElU/mL | NM | 72 ± 35 | 64 ± 34 |
| Albmg/L | 42 ± 3 | 41 ± 4 | 41 ± 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lundström, S.L.; Heyder, T.; Wiklundh, E.; Zhang, B.; Eklund, A.; Grunewald, J.; Zubarev, R.A. SpotLight Proteomics—A IgG-Enrichment Phenotype Profiling Approach with Clinical Implications. Int. J. Mol. Sci. 2019, 20, 2157. https://doi.org/10.3390/ijms20092157
Lundström SL, Heyder T, Wiklundh E, Zhang B, Eklund A, Grunewald J, Zubarev RA. SpotLight Proteomics—A IgG-Enrichment Phenotype Profiling Approach with Clinical Implications. International Journal of Molecular Sciences. 2019; 20(9):2157. https://doi.org/10.3390/ijms20092157
Chicago/Turabian StyleLundström, Susanna L., Tina Heyder, Emil Wiklundh, Bo Zhang, Anders Eklund, Johan Grunewald, and Roman A. Zubarev. 2019. "SpotLight Proteomics—A IgG-Enrichment Phenotype Profiling Approach with Clinical Implications" International Journal of Molecular Sciences 20, no. 9: 2157. https://doi.org/10.3390/ijms20092157





