Involvement of p38 Activation and Mitochondria in Death of Human Leukemia Cells Induced by an Agonistic Human Monoclonal Antibody Fab Specific to TRAIL Receptor 1
Abstract
:1. Introduction
2. Results
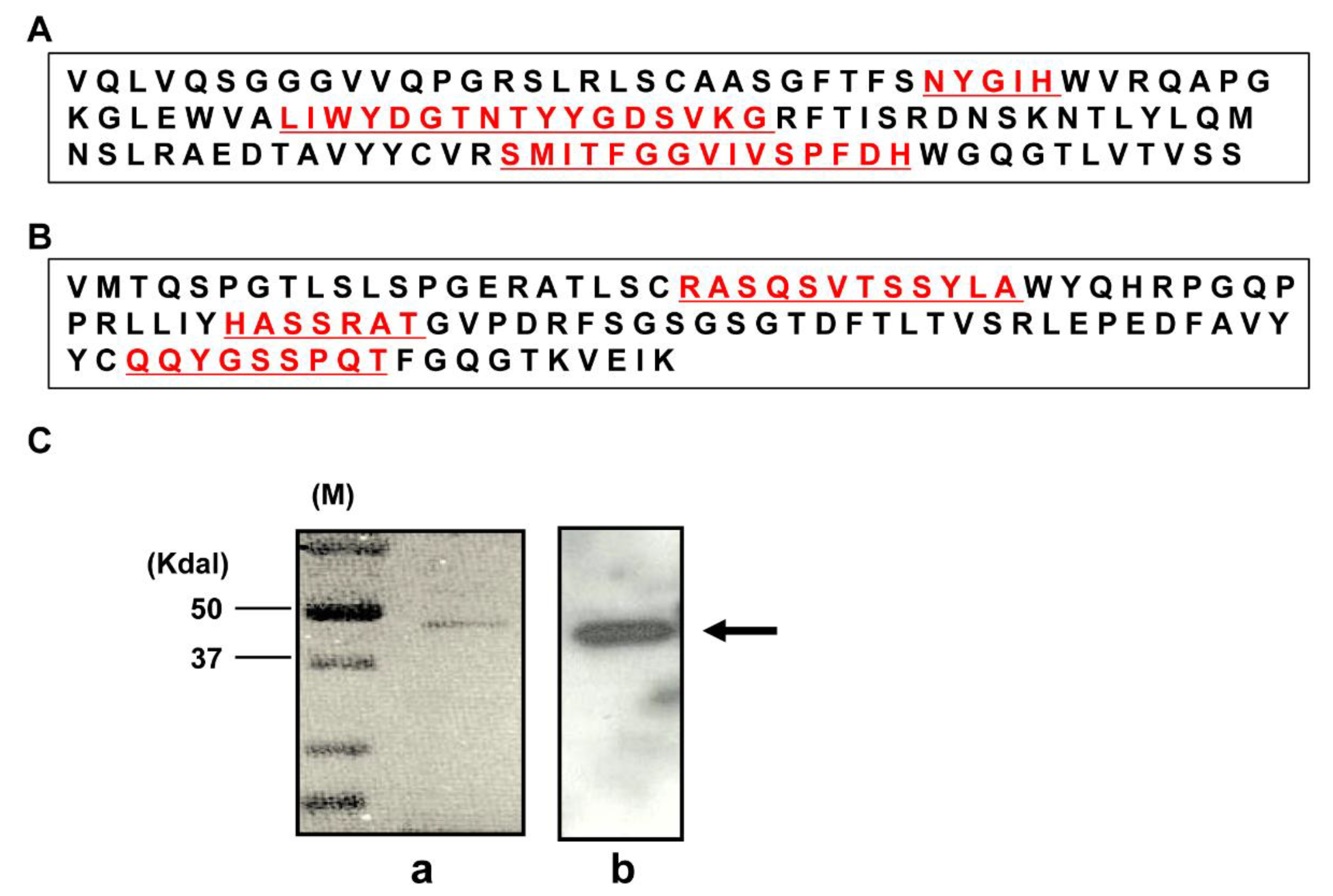
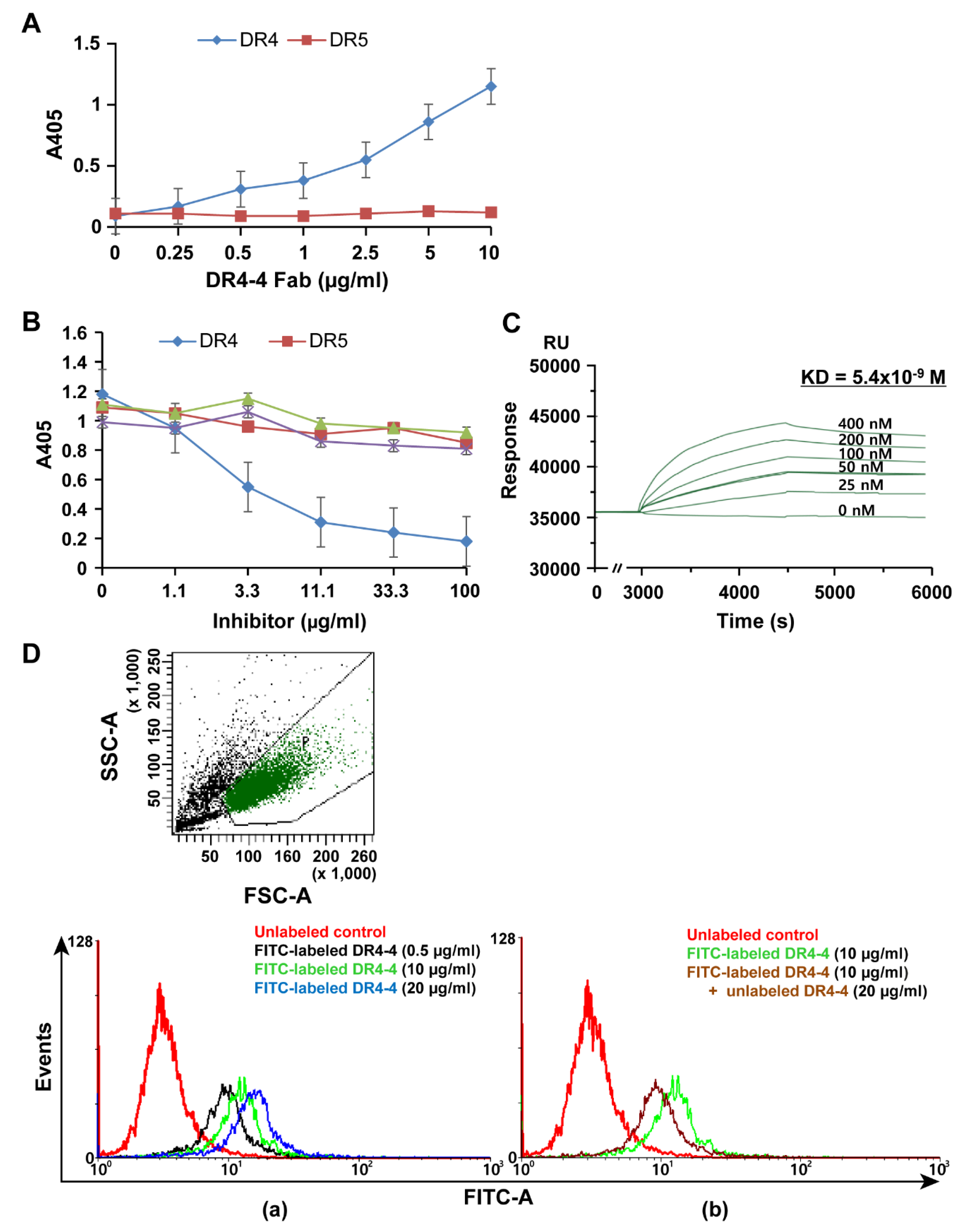
2.1. Characterization of a Human mAb Fab against DR4
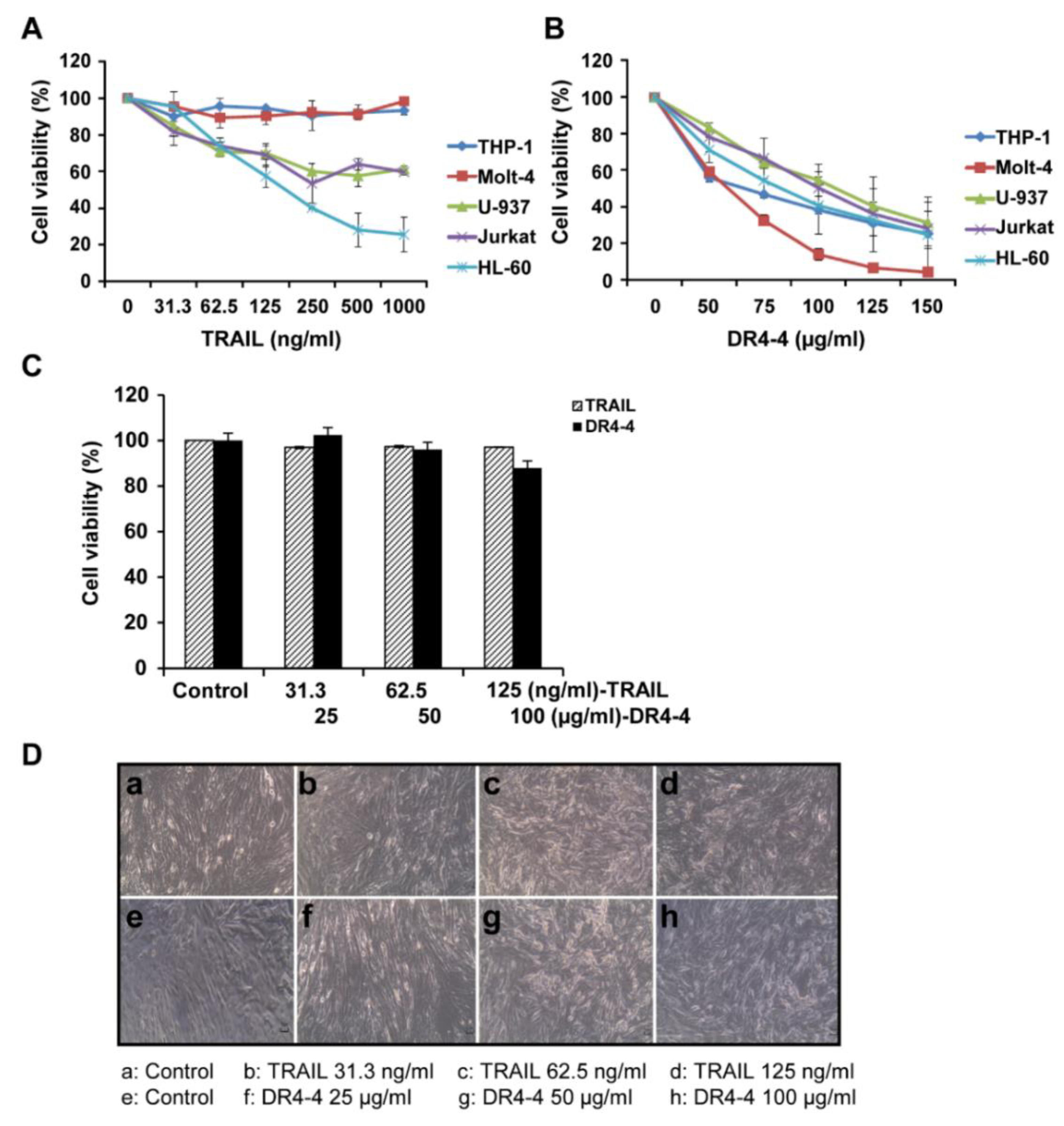
2.2. DR4-4 Induces Cell Death in Various Cancer Cells
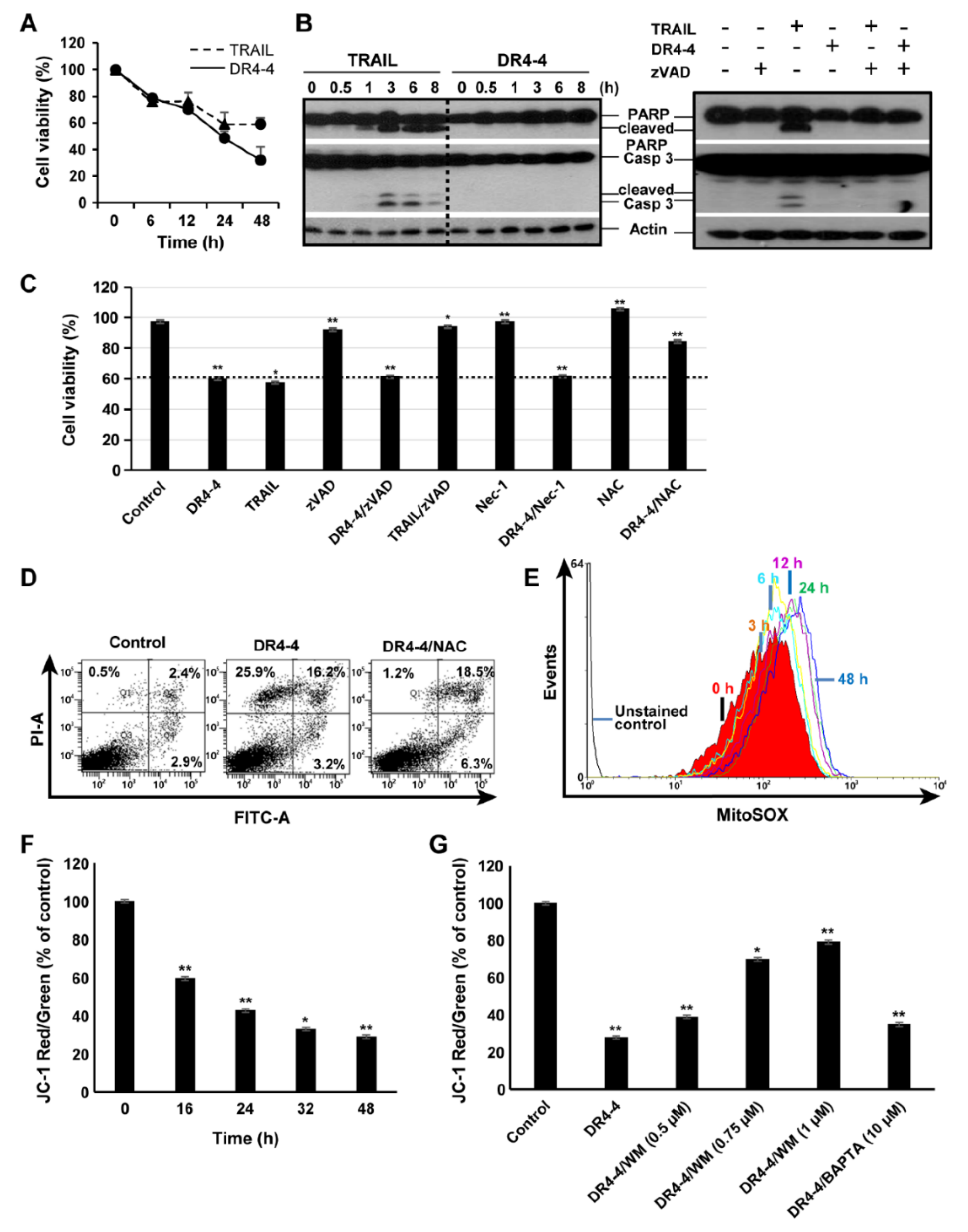
2.3. DR4-4 Fab Induces Caspase-Independent Cell Death
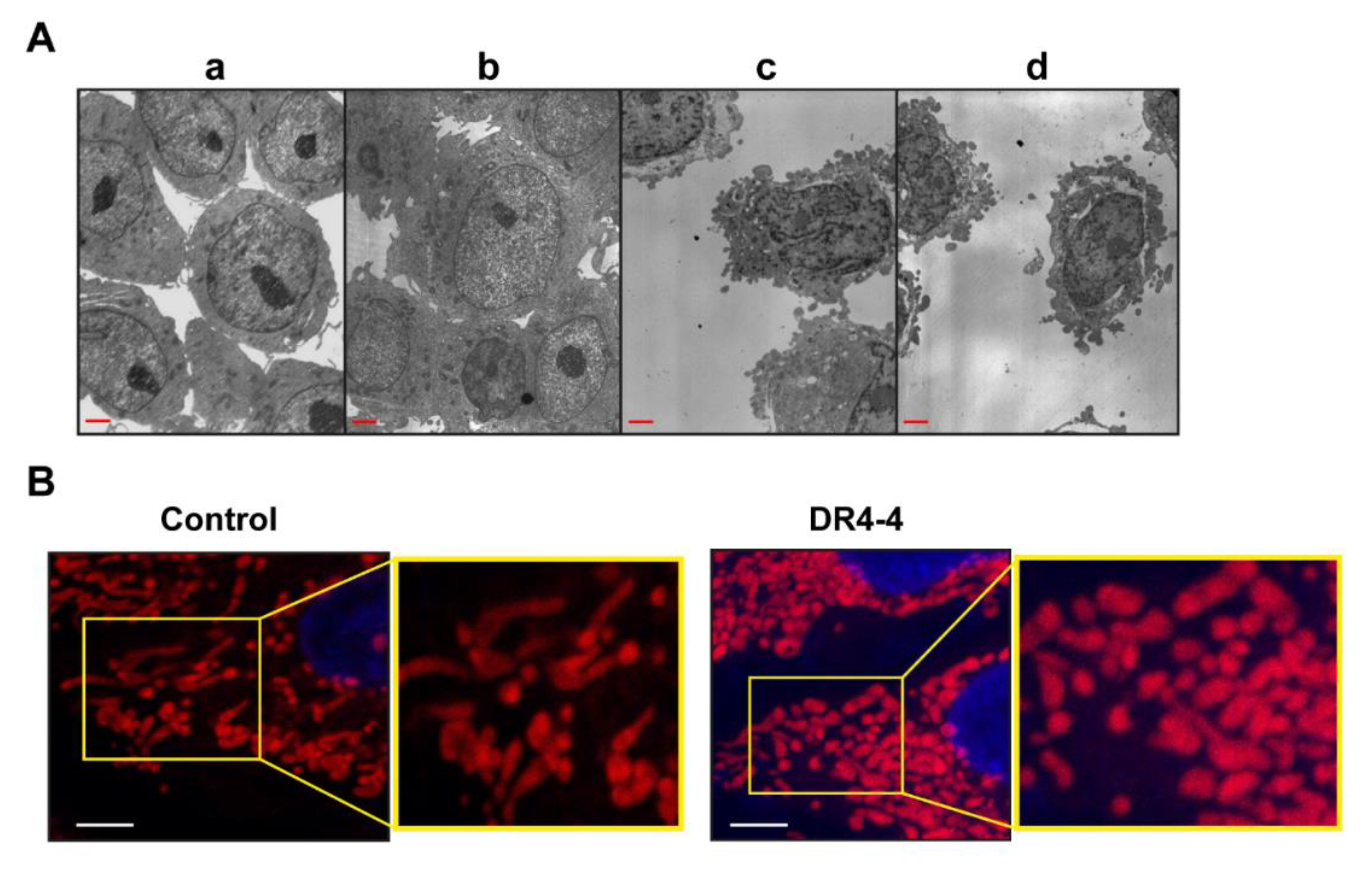
2.4. Mitochondria Are Involved in DR4-4 Fab–Induced Death
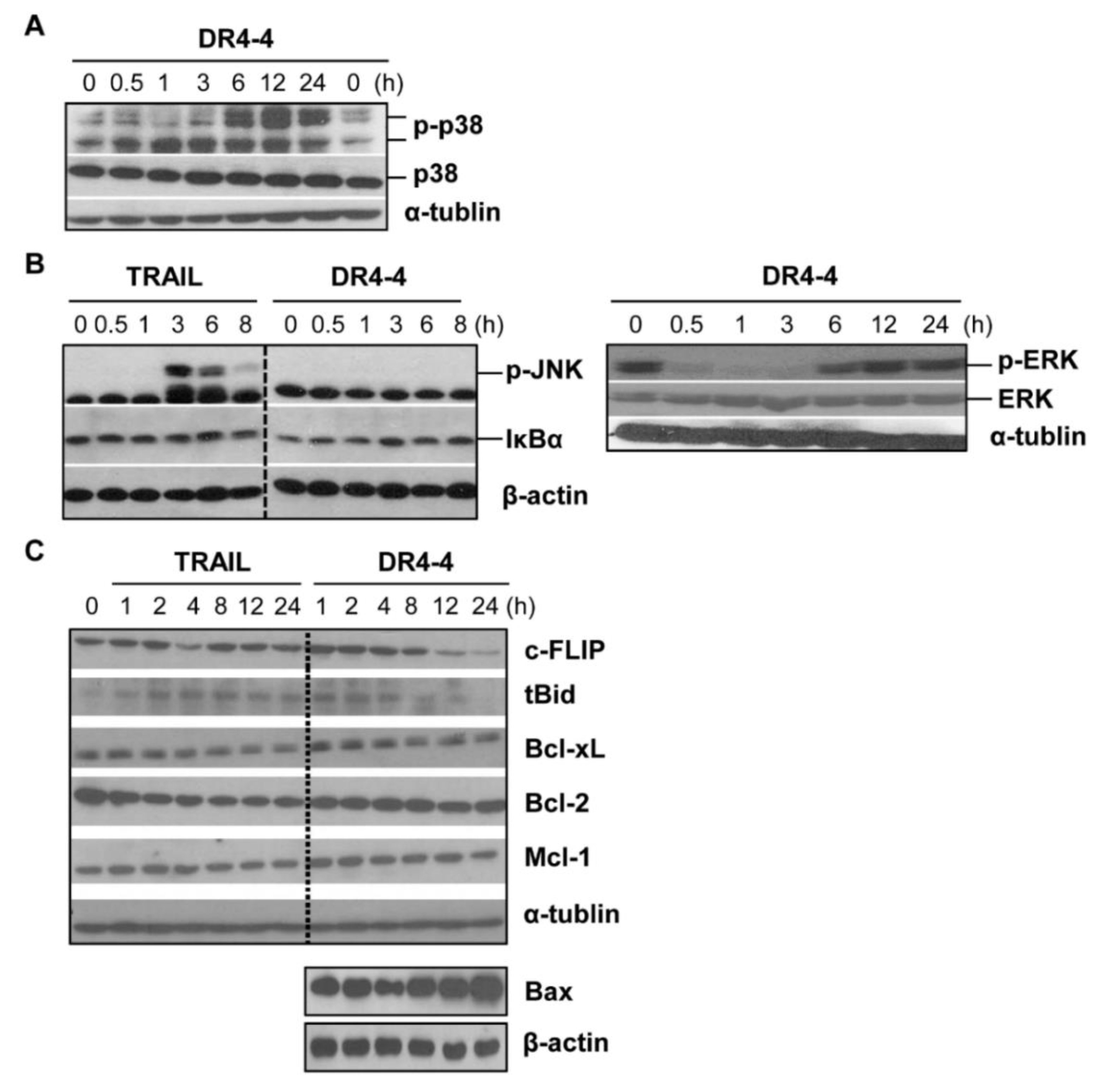
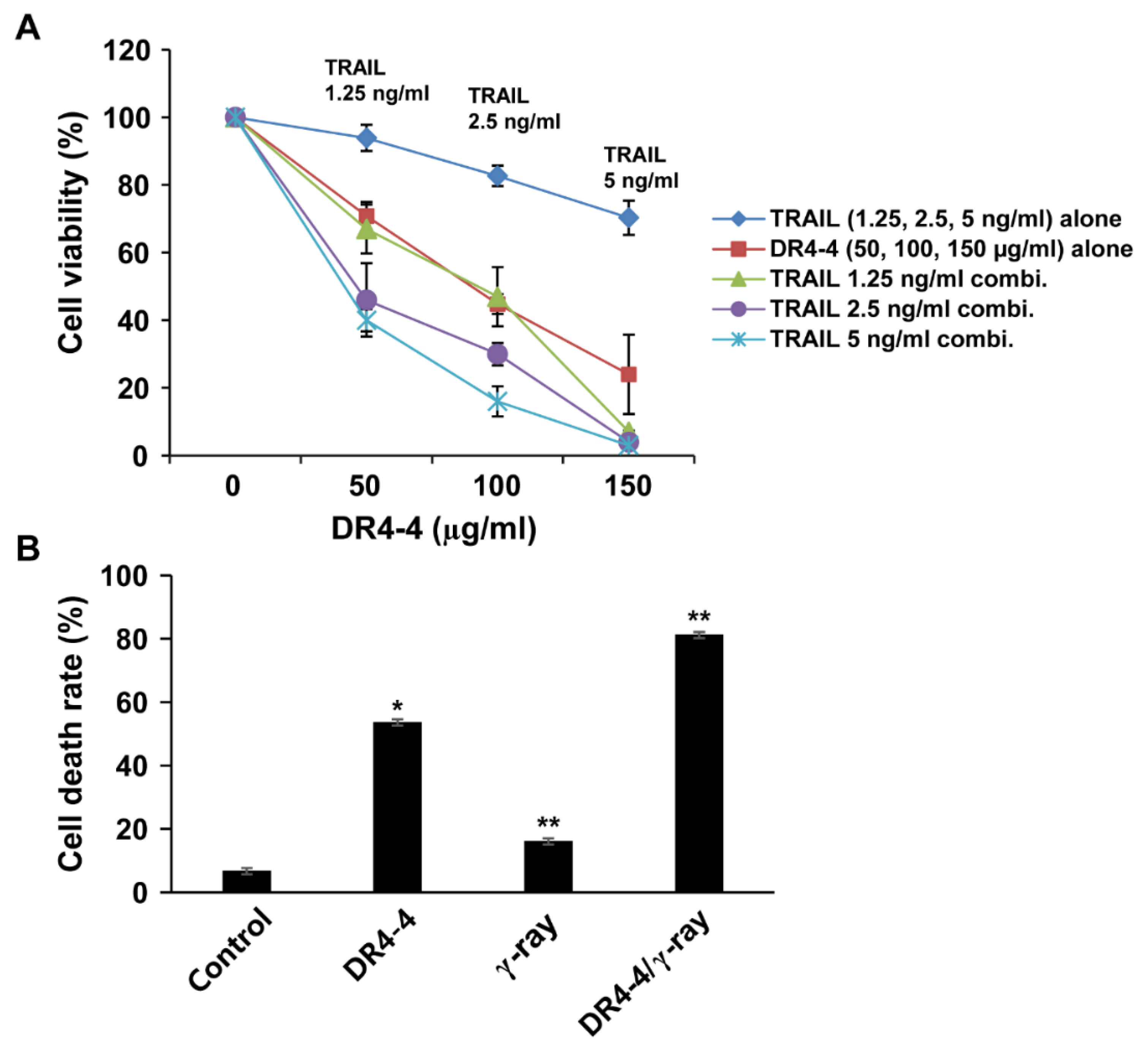
2.5. DR4-4 Fab Activates p38 MAPK, Inhibits c-FLIP, and Increases Cell Death Rates in Combination with TRAIL or γ-Irradiation
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Phage Display and Purification of Soluble Fab
4.3. Cell Culture
4.4. Immunoblotting
4.5. Direct Binding and Competitive ELISA
4.6. Affinity Analysis by SPR
4.7. FACS Analysis for Cellular Binding of DR4-4 Fab
4.8. Cytotoxicity Test
4.9. Mitochondrial ROS and MMP Measurement
4.10. Confocal Microscopy for Mitochondrial Morphology
4.11. Electron Microscopy for Cellular Image Analysis
4.12. γ-Irradiation
4.13. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Ab | antibody |
| BAPTA | 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid |
| Bcl-2 | B-cell lymphoma 2 |
| Bcl-xL | B-cell lymphoma extra-large |
| Bid | BH3 interacting-domain death agonist |
| c-FLIP | cellular FADD-like IL-1β-converting enzyme–inhibitory protein |
| DcR | decoy receptor |
| DISC | death-inducing signaling complex |
| DR | death receptor |
| ELISA | enzyme-linked immunosorbent assay |
| ERK | extracellular signal-regulated kinase |
| Fab | antigen binding fragment |
| FACS | fluorescence-activated cell sorting |
| FITC | fluorescein isothiocyanate |
| IκBα | nuclear factor κB inhibitor α |
| JNK | c-Jun N-terminal kinase |
| mAb | monoclonal antibody |
| Mcl-1 | myeloid cell leukemia 1 |
| MMP | mitochondrial membrane potential |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NAC | N-acetyl-l-cysteine |
| NF-κB | nuclear factor κB |
| p38 MAPK | p38 mitogen-activated protein kinase |
| PBS | phosphate-buffered saline |
| ROS | reactive oxygen species |
| SPR | surface plasmon resonance |
| sTRAIL | soluble TRAIL |
| tBid | truncated Bid |
| TNF | tumor necrosis factor |
| TRAIL | tumor necrosis factor–related apoptosis-inducing ligand |
| VH | heavy chain |
| VL | light chain |
| WM | wortmannin |
References
- Azijli, K.; Yuvaraj, S.; van Roosmalen, I.; Flach, K.; Giovannetti, E.; Peters, G.; de Jong, S.; Kruyt, F. MAPK p38 and JNK have opposing activities on TRAIL-induced apoptosis activation in NSCLC H460 cells that involves RIP1 and caspase-8 and is mediated by Mcl-1. Apoptosis 2013, 18, 851–860. [Google Scholar] [CrossRef]
- Bellail, A.; Qi, L.; Mulligan, P.; Chhabra, V.; Hao, C. TRAIL Agonists on Clinical Trials for Cancer Therapy: The Promises and the Challenges. Rev. Recent Clin. Trials 2009, 4, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-J.; Park, K.; Chung, H.; Kim, H. Analysis of the phenotypes of Jurkat clones with different TRAIL-sensitivities. Cancer Lett. 2003, 194, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Griffith, T.S.; Rauch, C.T.; Smolak, P.J.; Waugh, J.Y.; Boiani, N.; Lynch, D.H.; Smith, C.A.; Goodwin, R.G.; Kubin, M.Z. Functional analysis of TRAIL receptors using monoclonal antibodies. J. Immunol. 1999, 162, 2597–2605. [Google Scholar] [PubMed]
- Rowinsky, E.K. Targeted induction of apoptosis in cancer management: The emerging role of tumor necrosis factor-related apoptosis-inducing ligand receptor activating agents. J. Clin. Oncol. 2005, 23, 9394–9407. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Moosmayer, D.; Wuest, T.; Bartke, T.; Gerlach, E.; Schönherr, U.; Peters, N.; Scheurich, P.; Pfizenmaier, K. Differential activation of TRAIL-R1 and -2 by soluble and membrane TRAIL allows selective surface antigen-directed activation of TRAIL-R2 by a soluble TRAIL derivative. Oncogene 2001, 20, 4101–4106. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, D.; Shahrokh, Z.; Marsters, S.; Achilles, K.; Shih, D.; Mounho, B.; Hillan, K.; Totpal, K.; DeForge, L.; Schow, P.; et al. Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat. Med. 2001, 7, 383–385. [Google Scholar] [CrossRef]
- Walczak, H.; Miller, R.E.; Ariail, K.; Gliniak, B.; Griffith, T.S.; Kubin, M.; Chin, W.; Jones, J.; Woodward, A.; Le, T.; et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999, 5, 157–163. [Google Scholar] [CrossRef]
- Zauli, G.; Bosco, R.; Secchiero, P. Molecular targets for selective killing of TRAIL-resistant leukemic cells. Expert Opin. Therap. Targets 2011, 15, 931–942. [Google Scholar] [CrossRef]
- Degli-Esposti, M. To die or not to die-the quest of the TRAIL receptors. J. Leukocyte Biol. 1999, 65, 535–542. [Google Scholar] [CrossRef] [Green Version]
- Steele, L.P.; Georgopoulos, N.T.; Southgate, J.; Selby, P.J.; Trejdosiewicz, L.K. Differential susceptibility to TRAIL of normal versus malignant human urothelial cells. Cell Death Differ. 2006, 13, 1564–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jönsson, G.; Paulie, S.; Grandien, A. High level of cFLIP correlates with resistance to death receptor-induced apoptosis in bladder carcinoma cells. Anticancer Res. 2003, 23, 1213–1218. [Google Scholar] [PubMed]
- Okano, H.; Shiraki, K.; Inoue, H.; Kawakita, T.; Yamanaka, T.; Deguchi, M.; Sugimoto, K.; Sakai, T.; Ohmori, S.; Fujikawa, K.; et al. Cellular FLICE/caspase-8-inhibitory protein as a principal regulator of cell death and survival in human hepatocellular carcinoma. Lab Investig. 2003, 83, 1033–1043. [Google Scholar] [CrossRef]
- Weldon, C.B.; Parker, A.P.; Patten, D.; Elliott, S.; Tang, Y.; Frigo, D.E.; Dugan, C.M.; Coakley, E.L.; Butler, N.N.; Clayton, J.L.; et al. Sensitization of apoptotically-resistant breast carcinoma cells to TNF and TRAIL by inhibition of p38 mitogen-activated protein kinase signaling. Int. J. Oncol. 2004, 24, 1473–1480. [Google Scholar]
- Kim, Y.; Suh, N.; Sporn, M.; Reed, J.C. An Inducible Pathway for Degradation of FLIP Protein Sensitizes Tumor Cells to TRAIL-induced Apoptosis. J. Biol. Chem. 2002, 277, 22320–22329. [Google Scholar] [CrossRef] [Green Version]
- Cabeça, T.K.; de Mello Abreu, A.; Andrette, R.; de Souza Lino, V.; Morale, M.G.; Aguayo, F.; Termini, L.; Villa, L.L.; Lepique, A.P.; Boccardo, E. HPV-Mediated Resistance to TNF and TRAIL Is Characterized by Global Alterations in Apoptosis Regulatory Factors, Dysregulation of Death Receptors, and Induction of ROS/RNS. Int. J. Mol. Sci. 2019, 20, 198. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.R.; Reginato, M.; Debnath, J.; Queenan, B.; Brugge, J.S. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 3438–3443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alger, H.M.; Raben, N.; Pistilli, E.; Francia, D.L.; Rawat, R.; Getnet, D.; Ghimbovschi, S.; Chen, Y.W.; Lundberg, I.E.; Nagaraju, K. The Role of TRAIL in Mediating Autophagy in Myositis Skeletal Muscle. Arthr. Rheum. 2011, 63, 3448–3457. [Google Scholar] [CrossRef]
- Kemp, T.J.; Kim, J.S.; Crist, S.A.; Griffith, T.S. Induction of Necrotic Tumor Cell Death by TRAIL/Apo-2L. Apoptosis 2003, 8, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Meurette, O.; Rebillard, A.; Huc, L.; Le, M.G.; Merino, D.; Micheau, O.; Lagadic-Gossmann, D.; Dimanche-Boitre, M.-T. TRAIL Induces Receptor-Interacting Protein 1–Dependent and Caspase-Dependent Necrosis-Like Cell Death under Acidic Extracellular Conditions. Cancer Res. 2007, 67, 218–226. [Google Scholar] [CrossRef] [Green Version]
- Georgakis, G.V.; Li, Y.; Humphreys, R.; Andreeff, M.; O’Brien, S.; Younes, M.; Carbone, A.; Albert, V. Activity of selective fully human agonistic antibodies to the TRAIL death receptors TRAIL-R1 and TRAIL-R2 in primary and cultured lymphoma cells: Induction of apoptosis and enhancement of doxorubicin- and bortezomib-induced cell death. Brit. J. Haematol. 2005, 130, 501–510. [Google Scholar] [CrossRef]
- Motoki, K.; Mori, E.; Matsumoto, A.; Thomas, M.; Tomura, T.; Humphreys, R.; Albert, V.; Muto, M.; Yoshida, H.; Aoki, M.; et al. Enhanced apoptosis and tumor regression induced by a direct agonist antibody to tumor necrosis factor-related apoptosis-inducing ligand receptor 2. Clin. Cancer Res. 2005, 11, 3126–3135. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, C.; Zheng, Y.; Zhang, J.; Tao, X.; Liu, S.; Zheng, D.; Liu, Y. A novel anti-human DR5 monoclonal antibody with tumoricidal activity induces caspase-dependent and caspase-independent cell death. J. Biol. Chem. 2005, 280, 41940–41952. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, K.; Liu, W.; Zhao, L.; Wang, Z.; Liu, D.; Ohtsuka, T.; Zhang, H.; Mountz, J.D.; Koopman, W.J.; Kimberly, R.P.; et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat. Med. 2001, 7, 7954–7960. [Google Scholar] [CrossRef]
- Chuntharapai, A.; Dodge, K.; Grimmer, K.; Schroeder, K.; Marsters, S.A.; Koeppen, H.; Ashkenazi, A.; Kim, K.J. Isotype-dependent inhibition of tumor growth in vivo by monoclonal antibodies to death receptor 4. J. Immunol. 2001, 166, 4891–4898. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Yamaguchi, N.; Akiba, H.; Kojima, Y.; Hayakawa, Y.; Tanner, J.E.; Sayers, T.J.; Seki, N.; Okumura, K.; Yagita, H.; Smyth, M.J. Induction of tumor-specific T cell immunity by anti-DR5 antibody therapy. J. Exp. Med. 2004, 199, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Haynes, N.M.; Hawkins, E.D.; Li, M.; McLaughlin, N.M.; Hämmerling, G.J.; Schwendener, R.; Winoto, A.; Wensky, A.; Yagita, H.; Takeda, K.; et al. CD11c+ Dendritic Cells and B Cells Contribute to the Tumoricidal Activity of Anti-DR5 Antibody Therapy in Established Tumors. J. Immunol. 2010, 185, 532–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marjaneh, R.M.; Hassanian, S.M.; Ghobadi, N.; Ferns, G.A.; Karimi, A.; Jazayeri, M.H.; Nasiri, M.; Avan, A.; Khazaei, M. Targeting the death receptor signaling pathway as a potential therapeutic target in the treatment of colorectal cancer. J. Cell Physiol. 2018, 233, 6538–6549. [Google Scholar] [CrossRef]
- Menoret, E.; Gomez-Bougie, P.; Geffroy-Luseau, A.; Daniels, S.; Moreau, P.; Le, G.S.; Harousseau, J.L.; Bataille, R.; Amiot, M.; Pellat-Deceunynck, C. Mcl-1L cleavage is involved in TRAIL-R1- and TRAIL-R2-mediated apoptosis induced by HGS-ETR1 and HGS-ETR2 human mAbs in myeloma cells. Blood 2006, 108, 1346–1352. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Kim, S.Y.; Zhou, Z.; Lagasse, E.; Kwon, Y.T.; Lee, Y.J. Hyperthermia enhances mapatumumab-induced apoptotic death through ubiquitin-mediated degradation of cellular FLIP(long) in human colon cancer cells. Cell Death Dis. 2013, 4, 4. [Google Scholar] [CrossRef]
- Chen, L.; Qiu, Y.; Hao, Z.; Cai, J.; Zhang, S.; Liu, Y.; Zheng, D. A novel humanized anti-tumor necrosis factor-related apoptosis-inducing ligand-R2 monoclonal antibody induces apoptotic and autophagic cell death. IUBMB Life 2017, 69, 735–744. [Google Scholar] [CrossRef]
- Merchant, M.S.; Geller, J.I.; Baird, K.; Chou, A.J.; Galli, S.; Charles, A.; Amaoko, M.; Rhee, E.H.; Price, A.; Wexler, L.H. Phase I trial and pharmacokinetic study of lexatumumab in pediatric patients with solid tumors. J. Clin. Oncol. 2012, 30, 4141–4147. [Google Scholar] [CrossRef]
- Pukac, L.; Kanakaraj, P.; Humphreys, R.; Alderson, R.; Bloom, M.; Sung, C.; Riccobene, T.; Johnson, R.; Fiscella, M.; Mahoney, A.; et al. HGS-ETR1, a fully human TRAIL-receptor 1 monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br. J. Cancer 2005, 92, 1430–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oya, M.; Ohtsubo, M.; Takayanagi, A.; Tachibana, M.; Shimizu, N.; Murai, M. Constitutive activation of nuclear factor-kB prevents TRAIL-induced apoptosis in renal cancer cells. Oncogene 2001, 20, 3888–3896. [Google Scholar] [CrossRef] [PubMed]
- Keane, M.; Rubinstein, Y.; Cuello, M.; Ettenberg, S.; Banerjee, P.; Nau, M.; Lipkowitz, S. Inhibition of NF-κB activity enhances TRAIL mediated apoptosis in breast cancer cell lines. Breast Cancer Res. Treat. 2000, 64, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Xu, Z.; Myers, J.; Wu, X. Bypass NF-κB-mediated survival pathways by TRAIL and Smac. Cancer Biol. Ther. 2007, 6, 1031–1035. [Google Scholar] [CrossRef] [Green Version]
- Safa, A.R. Roles of c-FLIP in Apoptosis, Necroptosis, and Autophagy. J. Carcinog. Mutagen. 2013. [Google Scholar] [CrossRef]
- Naghdi, S.; Slovinsky, W.S.; Madesh, M.; Rubin, E.; Hajnóczky, G. Cell Death and Disease: a new journal for a central area of pathophysiology. Cell Death Dis. 2018, 9, 1028–1043. [Google Scholar] [CrossRef] [PubMed]
- Martın-Guerrero, S.M.; Muñoz-Gamez, J.A.; Carrasco, M.-C.; Salmeron, J.; Martın-Estebane, M.; Cuadros, M.A.; Navascues, J.; Martın-Oliva, D. Poly(ADP-ribose)polymerases inhibitors prevent early mitochondrial fragmentation and hepatocyte cell death induced by H2O2. PLoS ONE 2017, 12, 1–26. [Google Scholar] [CrossRef]
- Yang, Y.; Karakhanova, S.; Hartwig, W.; D’Haese, J.G.; Philippov, P.P.; Werner, J.; Bazhin, A.V. Mitochondria and mitochondrial ROS in cancer: Novel targets for anticancer therapy. J. Cell Physiol. 2016, 231, 2570–2581. [Google Scholar] [CrossRef]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Suresh, L.; Mehta, N.M.; Ram, R. Molecular targets in cerebral ischemia for developing novel therapeutics. Brain Res. Rev. 2007, 54, 34–66. [Google Scholar]
- Oh, H.-N.; Seo, J.-H.; Lee, M.-H.; Yoon, G.; Cho, S.-S.; Liu, K.; Choi, H.; Oh, K.B.; Cho, Y.S.; Kim, H. Oridonin induces apoptosis in oral squamous cell carcinoma probably through the generation of reactive oxygen species and the p38/JNK MAPK pathway. Int. J. Oncol. 2018, 52, 1749–1759. [Google Scholar] [CrossRef]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Kim, H.; Kim, S.; Basse, P.; Park, B.; Lee, B.; Lee, Y.J. Hyperthermia-enhanced TRAIL- and mapatumumab-induced apoptotic death is mediated through mitochondria in human colon cancer cells. J. Cell Biochem. 2012, 113, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xiao, F.; Li, Y.; Qian, W.; Ding, W.; Ye, X. Bypassing drug resistance by triggering necroptosis: Recent advances in mechanisms and its therapeutic exploitation in leukemia. J. Exp. Clin. Cancer Res. 2018, 37, 310–324. [Google Scholar] [CrossRef]
- Van Geelen, C.M.M.; Pennarun, B.; Boerma-Van, E.W.; Le, P.T.K.; Spierings, D.C.J.; de Vries, E.G.E.; de Jong, S. Downregulation of active caspase 8 as a mechanism of acquired TRAIL resistance in mismatch repair-proficient colon carcinoma cell lines. Int. J. Oncol. 2010, 37, 1031–1041. [Google Scholar]
- Micheau, O.; Thome, M.; Schneider, P.; Holler, N.; Tschopp, J.; Nicholson, DW.; Briand, C.; Grütter, MG. The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 2002, 277, 45162–45171. [Google Scholar] [CrossRef]
- Jeon, Y.E.; Seo, C.W.; Yu, E.S.; Lee, C.J.; Park, S.G.; Jang, Y.J. Characterization of human monoclonal autoantibody Fab fragments against oxidized LDL. Mol. Immunol. 2007, 44, 827–836. [Google Scholar] [CrossRef] [Green Version]
- Jang, Y.J.; Lecerf, J.M.; Stollar, B.D. Heavy chain dominance in the binding of DNA by a lupus mouse monoclonal autoantibody. Mol. Immunol. 1996, 33, 197–210. [Google Scholar] [CrossRef] [Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-R.; Hwang, E.; Jang, Y.-J. Involvement of p38 Activation and Mitochondria in Death of Human Leukemia Cells Induced by an Agonistic Human Monoclonal Antibody Fab Specific to TRAIL Receptor 1. Int. J. Mol. Sci. 2019, 20, 1967. https://doi.org/10.3390/ijms20081967
Lee Y-R, Hwang E, Jang Y-J. Involvement of p38 Activation and Mitochondria in Death of Human Leukemia Cells Induced by an Agonistic Human Monoclonal Antibody Fab Specific to TRAIL Receptor 1. International Journal of Molecular Sciences. 2019; 20(8):1967. https://doi.org/10.3390/ijms20081967
Chicago/Turabian StyleLee, You-Ri, Eunjoo Hwang, and Young-Ju Jang. 2019. "Involvement of p38 Activation and Mitochondria in Death of Human Leukemia Cells Induced by an Agonistic Human Monoclonal Antibody Fab Specific to TRAIL Receptor 1" International Journal of Molecular Sciences 20, no. 8: 1967. https://doi.org/10.3390/ijms20081967



