Roles of the Two-MnSOD System of Stenotrophomonas maltophilia in the Alleviation of Superoxide Stress
Abstract
:1. Introduction
2. Results
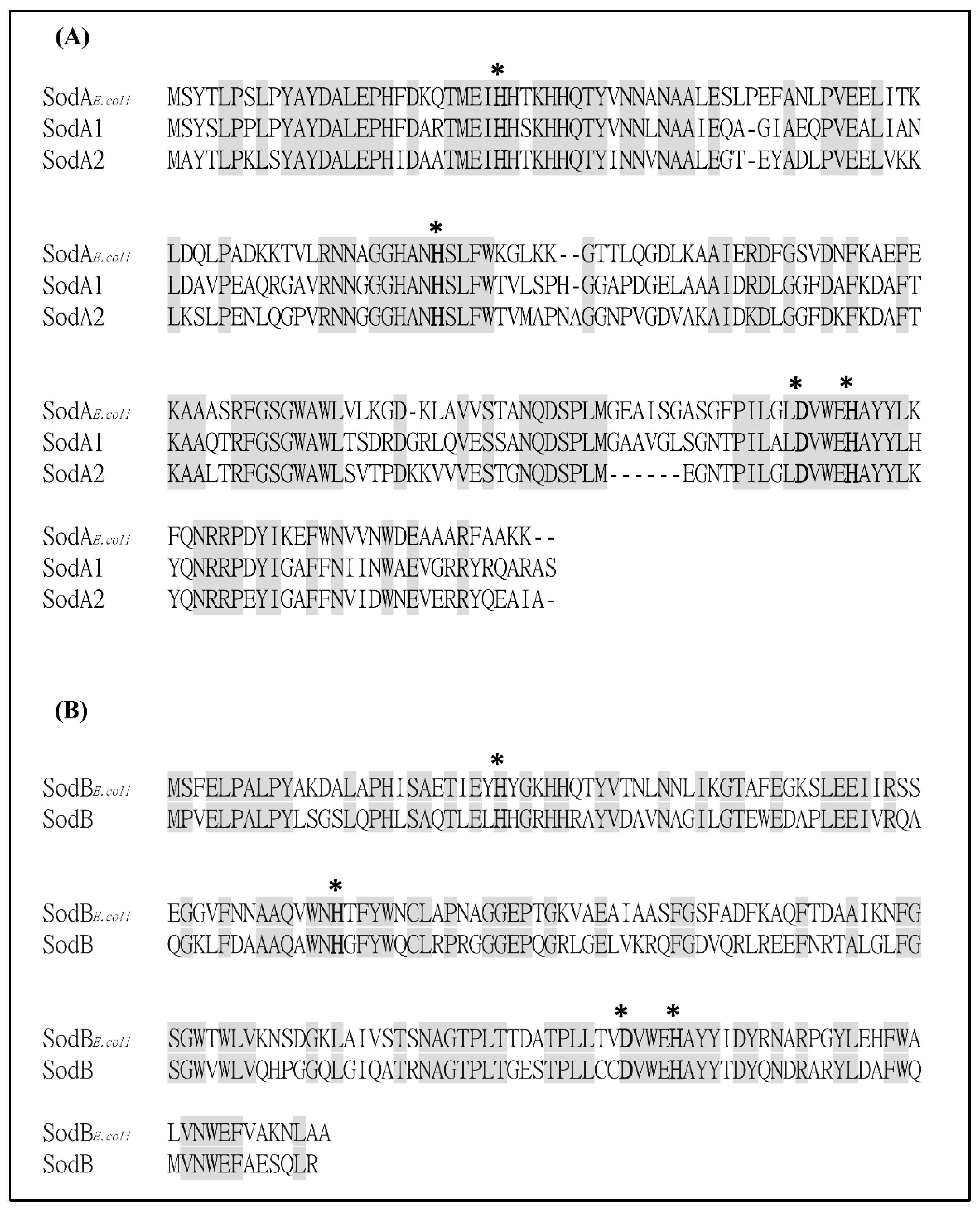
2.1. Bioinformatics Analysis of Putative SODs in S. maltophilia
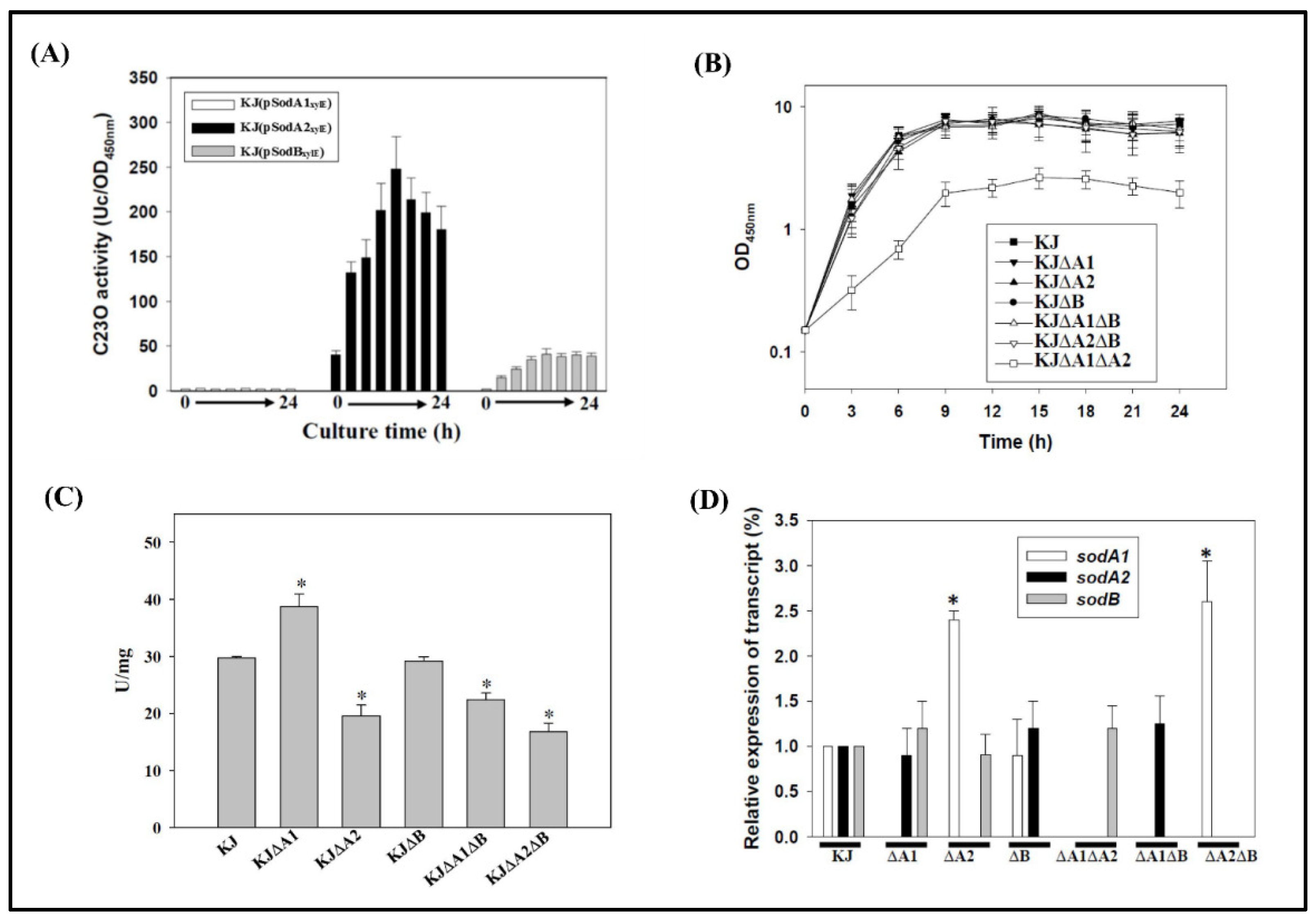
2.2. Implication of MnSODs and FeSOD in Aerobic Growth Conditions
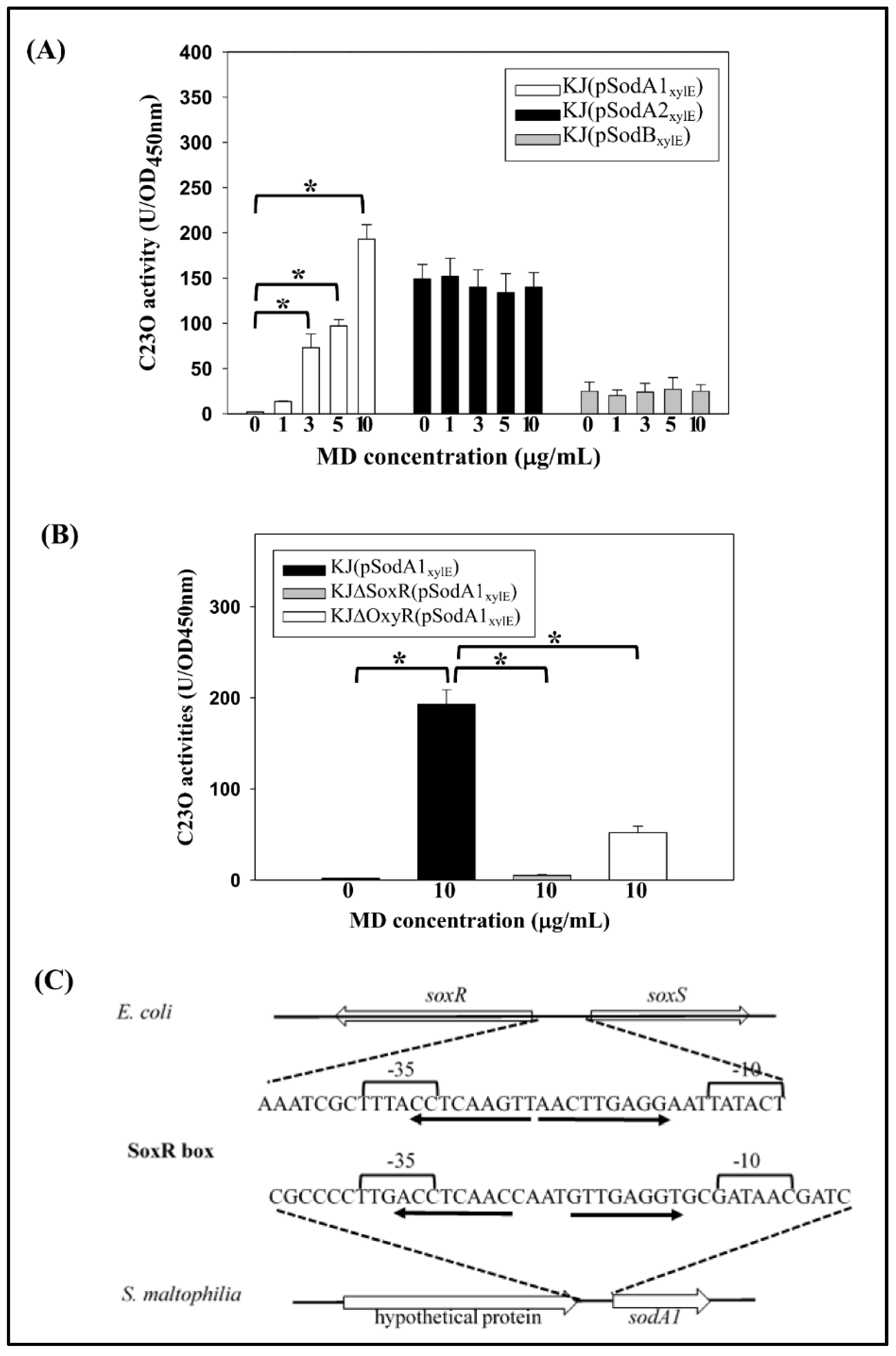
2.3. Expression of the Three SODs in Response to Menadione and Metal Stresses
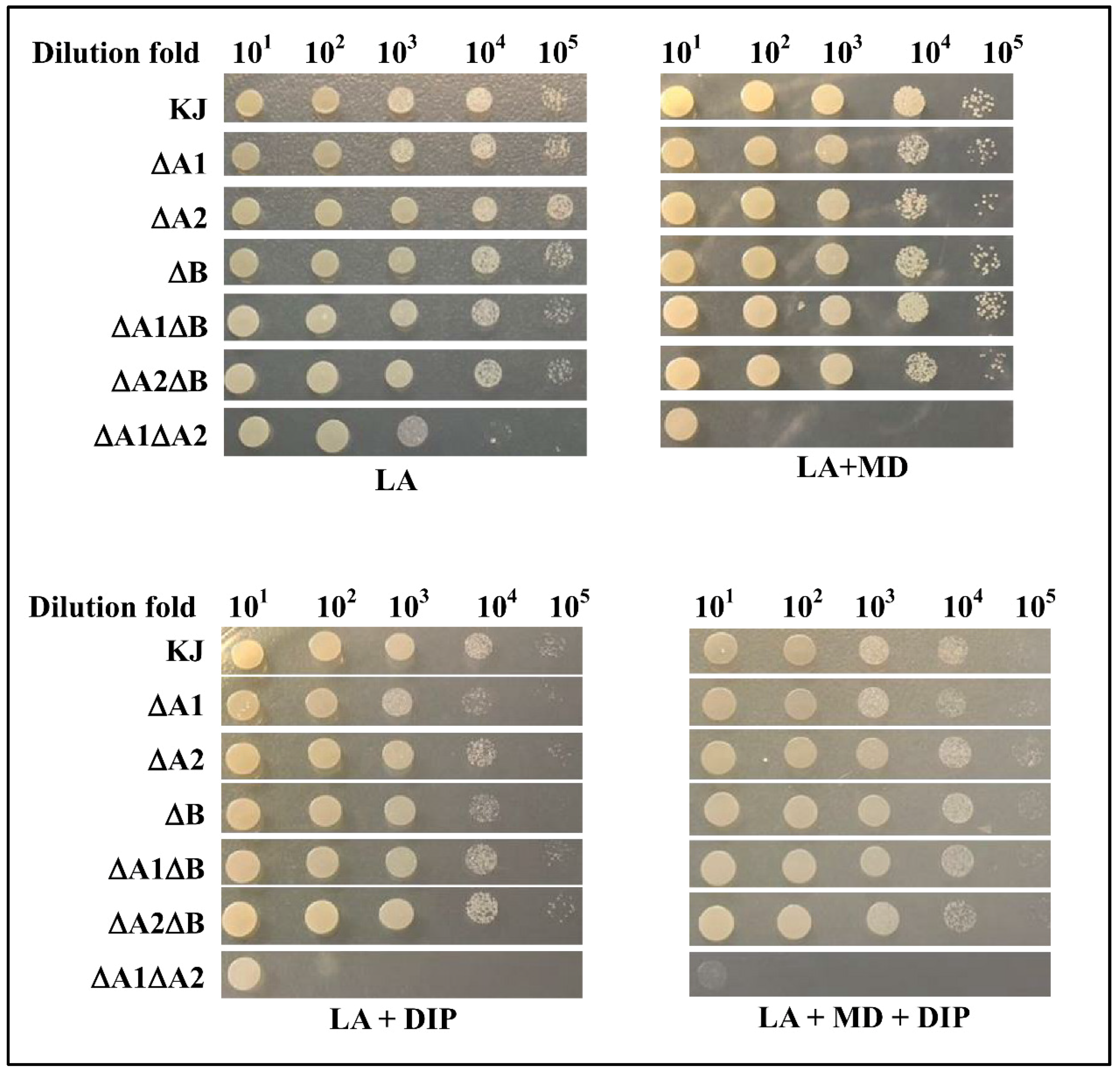
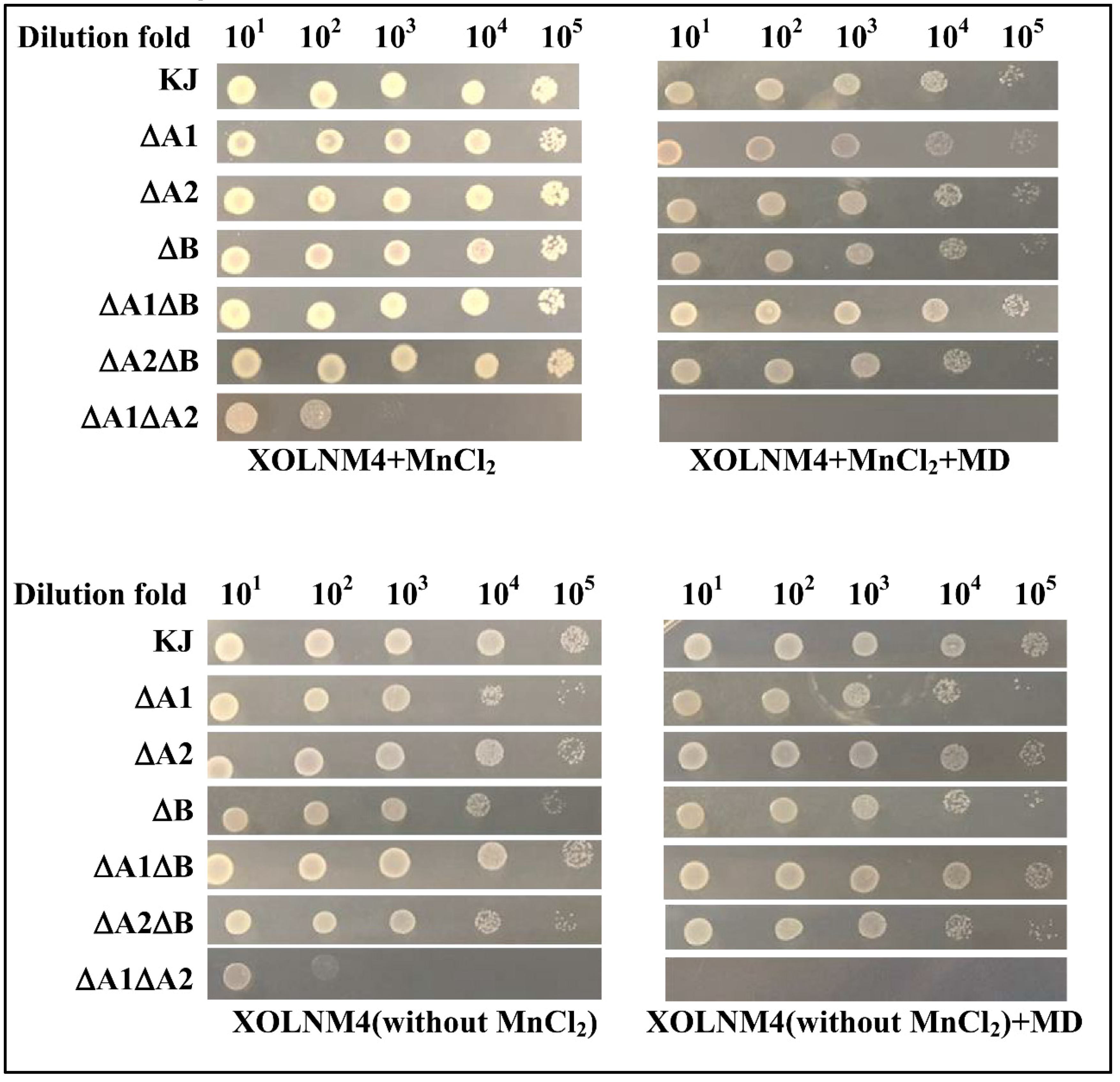
2.4. Implication of the Three SODs in Low-Iron and/or Superoxide Stress Conditions
2.5. Implication of the Three SODs in Low-Mn and/or Superoxide Stress Conditions
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmid, and Growth Condition
4.2. Construction of Deletion Mutants KJΔA1, KJΔA2, and KJΔB
4.3. SOD Activity Assay
4.4. Construction of Promoter–xylE Transcriptional Fusion Plasmids, pSodA1xylE, pSodA2xylE, and pSodBxylE
4.5. Determination of C23O Activity
4.6. Quantitative Real-Time PCR (qRT–PCR)
4.7. Cell Viability Assay
4.8. Sequence and Phylogenetic Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barnes, A.C.; Balebona, M.C.; Horne, M.T.; Ellis, A.E. Superoxide dismutase and catalase in Photobacterium damselae subsp. piscicida and their roles in resistance to reactive oxygen species. Microbiology 1999, 145, 483–494. [Google Scholar] [CrossRef]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 2–8. [Google Scholar]
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C 2011, 153, 175–190. [Google Scholar] [CrossRef]
- Miller, A.M. Superoxide dismutases: Ancient enzymes and new insights. FEBS Let. 2012, 586, 585–595. [Google Scholar] [CrossRef]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu. Rev. Bochem. 1995, 64, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Carlioz, A.; Touati, D. Isolation of superoxide dismutase mutants in Escherichia coli: Is superoxide dismutase necessary for aerobic life? EMBO J. 1986, 5, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Hassett, D.J.; Woodruff, W.A.; Wozniak, D.J.; Vasil, M.L.; Cohen, M.S.; Ohman, D.E. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: Demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J. Bacteriol. 1993, 175, 7658–7665. [Google Scholar] [CrossRef]
- Garcia, Y.M.; Barwinska-Sendra, A.; Tarrant, E.; Skaar, E.P.; Waldron, K.J.; Kehl-Fie, T.E. A superoxide dismutase capable of functioning with iron or manganese promotes the resistance of Staphylococcus aureus to calprotectin and nutritional immunity. PLoS Pathog. 2017, 13, e1006125. [Google Scholar] [CrossRef]
- Andrus, J.M.; Bowen, S.W.; Klaenhammer, T.R.; Hassan, H.M. Molecular characterization and functional analysis of the manganese-containing superoxide dismutase gene (sodA) from Streptococcus thermophilis A054. Arch. Biochem. Biophys. 2003, 420, 103–113. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Yang, C.H.; Wang, Q.; Mei, R. Two distinct manganese-containing superoxide dismutase genes in Bacillus cereus: Their physiological characterizations and roles in surviving in wheat rhizosphere. FEMS Microbiol. Lett. 2007, 272, 206–213. [Google Scholar] [CrossRef]
- Passalacqua, K.D.; Bergman, N.H.; Herring-Palmer, A.; Hanna, P. The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J. Bacteriol. 2006, 188, 3837–3848. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Gen. Biol. 2008, 9, R74. [Google Scholar] [CrossRef]
- Chiang, S.M.; Schellhorn, H.E. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 2012, 525, 161–169. [Google Scholar] [CrossRef]
- Kobayashi, K.; Tagawa, S. Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J. Biochem. 2004, 136, 607–615. [Google Scholar] [CrossRef]
- Kehl-Fie, T.E.; Skaar, E.P. Nutritional immunity beyond iron: A role for manganese and zinc. Curr. Opin. Chem. Biol. 2010, 14, 218–224. [Google Scholar] [CrossRef]
- Fu, H.; Yuan, J.; Gao, H. Microbial oxidative stress response: Novel insights from environmental facultative anaerobic bacteria. Arch. Biochem. Biophys. 2015, 584, 28–35. [Google Scholar] [CrossRef]
- Hassan, H.M.; Fridovich, I. Regulation of the synthesis of superoxide dismutase in Escherichia coli. Induction by methyl viologen. J. Biol. Chem. 1997, 252, 7667–7672. [Google Scholar]
- Hassett, D.J.; Schweizer, H.P.; Ohman, D.E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J. Bacteriol. 1995, 177, 6330–6337. [Google Scholar] [CrossRef]
- Polack, B.; Dacheux, D.; Delic-Attree, I.; Toussaint, B.; Vignais, P.M. Role of manganese superoxide dismutase in a mucoid isolate of Pseudomonas aeruginosa: Adaptation to oxidative stress. Infect. Immun. 1996, 64, 2216–2219. [Google Scholar]
- Chung, W.H. Unraveling new functions of superoxide dismutase using yeast model system: Beyond its conventional role in superoxide radical scavenging. J. Microbiol. 2017, 55, 409–416. [Google Scholar] [CrossRef]
- Pompilio, A.; Ciaardelli, D.; Crocetta, V.; Consalvo, A.; Zappacosta, R.; Di llio, C.; Di Bonaventura, G. Stenotrophomonas maltophilia virulence and specific variations in trace elements during acute lung infection: Implications in cystic fibrosis. PLoS ONE 2014, 9, e88769. [Google Scholar] [CrossRef]
- Huang, Y.W.; Huang, H.H.; Huang, K.H.; Chen, W.C.; Lin, Y.T.; Hsu, C.C.; Yang, T.C. AmpI functions as an iron exporter to alleviate β-lactam-mediated ROS stress in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2019, 63, e02467-18. [Google Scholar] [CrossRef]
- Yang, T.C.; Huang, Y.W.; Hu, R.M.; Huang, S.C.; Lin, Y.T. AmpDI is involved in expression of the chromosomal L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2009, 53, 2902–2907. [Google Scholar] [CrossRef]
- Hu, R.M.; Huang, K.J.; Wu, L.T.; Hsiao, Y.J.; Yang, T.C. Induction of L1 and L2 β-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2008, 52, 1198–1200. [Google Scholar] [CrossRef]
- Lin, C.W.; Huang, Y.W.; Hu, R.M.; Chiang, K.H.; Yang, T.C. The role of AmpR in regulation of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. Res. Microbiol. 2009, 160, 152–158. [Google Scholar] [CrossRef]
- Chen, C.H.; Huang, C.C.; Chung, T.C.; Hu, R.M.; Huang, Y.W.; Yang, T.C. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2011, 55, 5826–5833. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jair, H.-W.; Lu, H.-F.; Huang, Y.-W.; Pan, S.-Y.; Lin, I.-L.; Huang, H.-H.; Yang, T.-C. Roles of the Two-MnSOD System of Stenotrophomonas maltophilia in the Alleviation of Superoxide Stress. Int. J. Mol. Sci. 2019, 20, 1770. https://doi.org/10.3390/ijms20071770
Jair H-W, Lu H-F, Huang Y-W, Pan S-Y, Lin I-L, Huang H-H, Yang T-C. Roles of the Two-MnSOD System of Stenotrophomonas maltophilia in the Alleviation of Superoxide Stress. International Journal of Molecular Sciences. 2019; 20(7):1770. https://doi.org/10.3390/ijms20071770
Chicago/Turabian StyleJair, Herng-Woei, Hsu-Feng Lu, Yi-Wei Huang, Sz-Yun Pan, I-Ling Lin, Hsin-Hui Huang, and Tsuey-Ching Yang. 2019. "Roles of the Two-MnSOD System of Stenotrophomonas maltophilia in the Alleviation of Superoxide Stress" International Journal of Molecular Sciences 20, no. 7: 1770. https://doi.org/10.3390/ijms20071770



