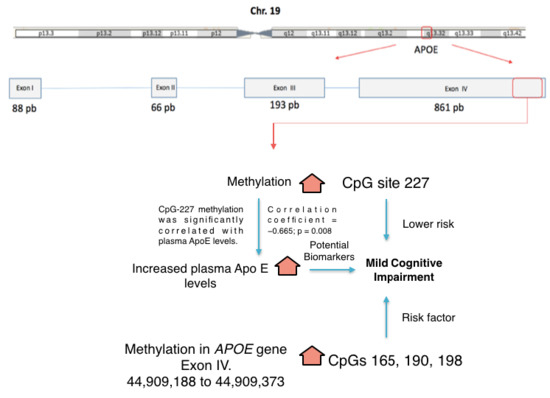
Differential Methylation in APOE (Chr19; Exon Four; from 44,909,188 to 44,909,373/hg38) and Increased Apolipoprotein E Plasma Levels in Subjects with Mild Cognitive Impairment
Abstract
:1. Introduction
2. Results
2.1. Baseline Characteristics
2.2. Plasma ApoE Levels and APOE Methylation
2.3. Plasma ApoE Levels and APOE Methylation Levels as Risk Factors for MCI
2.4. Correlation between Plasma ApoE Levels and APOE Methylation Levels
3. Discussion
4. Materials and Methods
4.1. Study Design and Population Sample
4.2. Medical Evaluation
4.3. Neuropsychological (NP) Evaluation
4.4. Diagnostic Classification of the Participants
4.4.1. Mild Cognitive Impairment
4.4.2. Normal Performance in Healthy Subjects
4.5. DNA Extraction and Bisulfite Treatment
4.6. Bisulfite Sequencing PCR (BSP)
4.7. Apolipoprotein Plasma Levels
4.8. Statistical Analysis
5. Limitations
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gauthier, S.; Reisberg, B.; Zaudig, M.; Petersen, R.C.; Ritchie, K.; Broich, K.; Belleville, S.; Brodaty, H.; Bennett, D.; Chertkow, H. Mild cognitive impairment. Lancet 2006, 367, 1262–1270. [Google Scholar] [CrossRef]
- Henao-Arboleda, E.; Aguirre-Acevedo, D.; Munoz, C.; Pineda, D.; Lopera, F. Prevalence of mild cognitive impairment, amnestic-type, in a Colombian population. Rev. Neurol. 2008, 46, 709–713. [Google Scholar] [PubMed]
- Hänninen, T.; Hallikainen, M.; Tuomainen, S.; Vanhanen, M.; Soininen, H. Prevalence of mild cognitive impairment: A population-based study in elderly subjects. Acta Neurol. Scand. 2002, 106, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C.; Roberts, R.O.; Knopman, D.S.; Boeve, B.F.; Geda, Y.E.; Ivnik, R.J.; Smith, G.E.; Jack, C.R. Mild cognitive impairment: Ten years later. Arch. Neurol. 2009, 66, 1447–1455. [Google Scholar] [CrossRef]
- Manly, J.J.; Tang, M.X.; Schupf, N.; Stern, Y.; Vonsattel, J.P.G.; Mayeux, R. Frequency and course of mild cognitive impairment in a multiethnic community. Ann. Neurol. 2008, 63, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Kaduszkiewicz, H.; Eisele, M.; Wiese, B.; Prokein, J.; Luppa, M.; Luck, T.; Jessen, F.; Bickel, H.; Mösch, E.; Pentzek, M. Prognosis of mild cognitive impairment in general practice: Results of the German AgeCoDe study. Ann. Family Med. 2014, 12, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; Van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: A meta-analysis. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Maestre, G.E.; Mena, L.J.; Melgarejo, J.D.; Aguirre-Acevedo, D.C.; Pino-Ramírez, G.; Urribarrí, M.; Chacon, I.J.; Chávez, C.A.; Falque-Madrid, L.; Gaona, C.A. Incidence of dementia in elderly Latin Americans: Results of the Maracaibo Aging Study. Alzheimer’s Dement. 2018, 14, 140–147. [Google Scholar] [CrossRef]
- Petersen, R.C.; Roberts, R.O.; Knopman, D.S.; Geda, Y.E.; Cha, R.H.; Pankratz, V.; Boeve, B.; Tangalos, E.; Ivnik, R.; Rocca, W. Prevalence of mild cognitive impairment is higher in men The Mayo Clinic Study of Aging. Neurology 2010, 75, 889–897. [Google Scholar] [CrossRef]
- Plassman, B.L.; Langa, K.M.; Fisher, G.G.; Heeringa, S.G.; Weir, D.R.; Ofstedal, M.B.; Burke, J.R.; Hurd, M.D.; Potter, G.G.; Rodgers, W.L. Prevalence of cognitive impairment without dementia in the United States. Ann. Intern. Med. 2008, 148, 427–434. [Google Scholar] [CrossRef]
- Sosa, A.L.; Albanese, E.; Stephan, B.C.; Dewey, M.; Acosta, D.; Ferri, C.P.; Guerra, M.; Huang, Y.; Jacob, K.; Jimenez-Velazquez, I.Z. Prevalence, distribution, and impact of mild cognitive impairment in Latin America, China, and India: A 10/66 population-based study. PLoS Med. 2012, 9, e1001170. [Google Scholar] [CrossRef] [PubMed]
- Schargrodsky, H.; Hernández-Hernández, R.; Champagne, B.M.; Silva, H.; Vinueza, R.; Ayçaguer, L.C.S.; Touboul, P.-J.; Boissonnet, C.P.; Escobedo, J.; Pellegrini, F. CARMELA: Assessment of cardiovascular risk in seven Latin American cities. Am. J. Med. 2008, 121, 58–65. [Google Scholar] [CrossRef]
- Melgarejo, J.D.; Maestre, G.E.; Thijs, L.; Asayama, K.; Boggia, J.; Casiglia, E.; Hansen, T.W.; Imai, Y.; Jacobs, L.; Jeppesen, J. Prevalence, treatment, and control rates of conventional and ambulatory hypertension across 10 populations in 3 continents. Hypertension 2017, 70, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Knopman, D.; Boland, L.; Mosley, T.; Howard, G.; Liao, D.; Szklo, M.; McGovern, P.; Folsom, A. Atherosclerosis Risk in Communities (ARIC) Study Investigators. Cardiovascular risk factors and cognitive decline in middle-aged adults. Neurology 2001, 56, 42–48. [Google Scholar] [CrossRef]
- Kivipelto, M.; Ngandu, T.; Fratiglioni, L.; Viitanen, M.; Kåreholt, I.; Winblad, B.; Helkala, E.-L.; Tuomilehto, J.; Soininen, H.; Nissinen, A. Obesity and vascular risk factors at midlife and the risk of dementia and Alzheimer disease. Arch. Neurol. 2005, 62, 1556–1560. [Google Scholar] [CrossRef] [PubMed]
- Román, G.C.; Mancera-Páez, O.; Bernal, C. Epigenetic Factors in Late-Onset Alzheimer’s disease: MTHFR and CTH Gene Polymorphisms, Metabolic Trans-sulfuration and Methylation Pathways, and B Vitamins. Int. J. Mol. Sci. 2019, 20, 319. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.-X.; Stern, Y.; Marder, K.; Bell, K.; Gurland, B.; Lantigua, R.; Andrews, H.; Feng, L.; Tycko, B.; Mayeux, R. The APOE-ε4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA 1998, 279, 751–755. [Google Scholar] [CrossRef]
- Taddei, K.; Clarnette, R.; Gandy, S.E.; Martins, R.N. Increased plasma apolipoprotein E (apoE) levels in Alzheimer’s disease. Neurosci. Lett. 1997, 223, 29–32. [Google Scholar] [CrossRef]
- Sullivan, P.; Han, B.; Liu, F.; Mace, B.; Ervin, J.; Wu, S.; Koger, D.; Paul, S.; Bales, K. Reduced levels of human apoE4 protein in an animal model of cognitive impairment. Neurobiol. Aging 2011, 32, 791–801. [Google Scholar] [CrossRef]
- Song, F.; Poljak, A.; Crawford, J.; Kochan, N.A.; Wen, W.; Cameron, B.; Lux, O.; Brodaty, H.; Mather, K.; Smythe, G.A. Plasma apolipoprotein levels are associated with cognitive status and decline in a community cohort of older individuals. PLoS ONE 2012, 7, e34078. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.; Roses, A.D.; Haines, J.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, K.; Zhao, H. Haplotype-association analysis. Adv. Genet. 2008, 60, 335–405. [Google Scholar] [PubMed]
- Chartier-Hariln, M.-C.; Parfitt, M.; Legrain, S.; Pérez-Tur, J.; Brousseau, T.; Evans, A.; Berr, C.; Vldal, O.; Roques, P.; Gourlet, V. Apolipoprotein E, ε4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: Analysis of the 19q13. 2 chromosomal region. Hum. Mol. Genet. 1994, 3, 569–574. [Google Scholar] [CrossRef]
- Ciceri, F.; Rotllant, D.; Maes, T. Understanding epigenetic alterations in Alzheimer’s and Parkinson’s disease: Towards targeted biomarkers and therapies. Curr. Pharm. Des. 2017, 23, 839–857. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.G.; Kim, J.Y.; Choi, J.K. Frequent hypermethylation of orphan CpG islands with enhancer activity in cancer. BMC Med. Genomics 2016, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Shaw, M.; Todd, K.; Khrestian, M.; D’Aleo, G.; Barnard, P.J.; Zahratka, J.; Pillai, J.; Yu, C.-E.; Keene, C.D. DNA methylation of TOMM40-APOE-APOC2 in Alzheimer’s disease. J. Hum. Genet. 2018, 63, 459. [Google Scholar] [CrossRef] [PubMed]
- Foraker, J.; Millard, S.P.; Leong, L.; Thomson, Z.; Chen, S.; Keene, C.D.; Bekris, L.M.; Yu, C.-E. The APOE gene is differentially methylated in Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 48, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.L.; Tybjærg-Hansen, A.; Nordestgaard, B.G.; Frikke-Schmidt, R. Plasma levels of apolipoprotein E and risk of dementia in the general population. Ann. Neurol. 2015, 77, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Paul, L.; Cattaneo, M.; D’Angelo, A.; Sampietro, F.; Fermo, I.; Razzari, C.; Fontana, G. Telomere length in peripheral blood mononuclear cells is associated with folate status in men in me. J. Nutr. 2009, 139, 1273–1278. [Google Scholar] [CrossRef]
- Cramer, P.E.; Cirrito, J.R.; Wesson, D.W.; Lee, C.D.; Karlo, J.C.; Zinn, A.E.; Casali, B.T.; Restivo, J.L.; Goebel, W.D.; James, M.J. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science 2012, 335, 1503–1506. [Google Scholar] [CrossRef]
- Larsen, P.A.; Lutz, M.W.; Hunnicutt, K.E.; Mihovilovic, M.; Saunders, A.M.; Yoder, A.D.; Roses, A.D. The Alu neurodegeneration hypothesis: A primate-specific mechanism for neuronal transcription noise, mitochondrial dysfunction, and manifestation of neurodegenerative disease. Alzheimer’s Dement. 2017, 13, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhao, W.; Ware, E.B.; Turner, S.T.; Mosley, T.H.; Smith, J.A. DNA methylation in the APOE genomic region is associated with cognitive function in African Americans. BMC Med. Genomics 2018, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Hernández, H.G.; Mahecha, M.F.; Mejía, A.; Arboleda, H.; Forero, D.A. Global long interspersed nuclear element 1 DNA methylation in a Colombian sample of patients with late-onset Alzheimer’s disease. Am. J. Alzheimer’s Dis. Other Dement. 2014, 29, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Du, A.; Jahng, G.; Hayasaka, S.; Kramer, J.; Rosen, H.; Gorno-Tempini, M.; Rankin, K.; Miller, B.; Weiner, M.; Schuff, N. Hypoperfusion in frontotemporal dementia and Alzheimer disease by arterial spin labeling MRI. Neurology 2006, 67, 1215–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosun, D.; Schuff, N.; Jagust, W.; Weiner, M.W.; Initiative, A.s.D.N. Discriminative power of arterial spin labeling magnetic resonance imaging and 18F-fluorodeoxyglucose positron emission tomography changes for amyloid-β-positive subjects in the Alzheimer’s disease continuum. Neurodegener. Dis. 2016, 16, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Fällmar, D.; Haller, S.; Lilja, J.; Danfors, T.; Kilander, L.; Tolboom, N.; Egger, K.; Kellner, E.; Croon, P.M.; Verfaillie, S.C. Arterial spin labeling-based Z-maps have high specificity and positive predictive value for neurodegenerative dementia compared to FDG-PET. Eur. Radiol. 2017, 27, 4237–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musiek, E.S.; Chen, Y.; Korczykowski, M.; Saboury, B.; Martinez, P.M.; Reddin, J.S.; Alavi, A.; Kimberg, D.Y.; Wolk, D.A.; Julin, P. Direct comparison of fluorodeoxyglucose positron emission tomography and arterial spin labeling magnetic resonance imaging in Alzheimer’s disease. Alzheimer’s Dement. 2012, 8, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Wolk, D.A.; Detre, J.A. Arterial spin labeling MRI: An emerging biomarker for Alzheimer’s disease and other neurodegenerative conditions. Curr. Opin. Neurol. 2012, 25, 421. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-E.; Cudaback, E.; Foraker, J.; Thomson, Z.; Leong, L.; Lutz, F.; Gill, J.A.; Saxton, A.; Kraemer, B.; Navas, P.; et al. Epigenetic signature and enhancer activity of the human APOE gene. Hum. Mol. Genet. 2013, 22, 5036–5047. [Google Scholar] [CrossRef] [Green Version]
- Estrada-Orozco, K.; Bonilla-Vargas, K.; Cruz, F.; Mancera, O.; Ruiz, M.; Alvarez, L.; Pardo, R.; Arboleda, H. Cognitive Assessment Test: Validation of a Short Cognitive Test for the Detection of Mild Cognitive Disorder. Int. J. Alzheimer’s Dis. 2018, 2018, 3280621. [Google Scholar] [CrossRef]
- Espitia Mendieta, D. Funciones Ejecutivas en el Envejecimiento Normal: Datos Normativos Con La Batería Neuronorma. Master’s Thesis, Universidad Nacional de Colombia, Bogotá, Colombia, 2017. [Google Scholar]
- Peña-Casanova, J.; Quiñones-Ubeda, S.; Gramunt-Fombuena, N.; Quintana-Aparicio, M.; Aguilar, M.; Badenes, D.; Cerulla, N.; Molinuevo, J.L.; Ruiz, E.; Robles, A.; et al. Spanish Multicenter Normative Studies (NEURONORMA Project): Norms for verbal fluency tests. Arch. Clin. Neuropsychol. 2009, 24, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Benavides, G.; Peña-Casanova, J.; Casals-Coll, M.; Gramunt, N.; Manero, R.M.; Puig-Pijoan, A.; Aguilar, M.; Robles, A.; Antúnez, C.; Frank-García, A.; et al. One-Year Reference Norms of Cognitive Change in Spanish Old Adults: Data from the NEURONORMA Sample. Arch. Clin. Neuropsychol. 2016, 31, 378–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranciva, F.; Casals-Coll, M.; Sánchez-Benavides, G.; Quintana, M.; Manero, R.M.; Rognoni, T.; Calvo, L.; Palomo, R.; Tamayo, F.; Peña-Casanova, J. Spanish normative studies in a young adult population (NEURONORMA young adults Project): Norms for the Boston Naming Test and the Token Test. Neurología 2012, 27, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Peña-Casanova, J.; Quiñones-Ubeda, S.; Gramunt-Fombuena, N.; Quintana, M.; Aguilar, M.; Molinuevo, J.L.; Serradell, M.; Robles, A.; Barquero, M.S.; Payno, M.; et al. Spanish Multicenter Normative Studies (NEURONORMA Project): Norms for the Stroop color-word interference test and the Tower of London-Drexel. Arch. Clin. Neuropsychol. 2009, 24, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Sanabria, F.; Bonilla-Vargas, K.; Estrada, K.; Mancera, O.; Vega, E.; Guerrero, E.; Ortega-Rojas, J.; Mahecha María, F.; Romero, A.; Montañés, P.; et al. Análisis de desempeños cognitivos y polimorfismos en SORL, PVRL2, CR1, TOMM40, APOE, PICALM, GWAS_14q, CLU y BIN1 en pacientes con trastorno neurocognitivo leve y en sujetos cognitivamente sanos. Neurología 2018. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, C.A.C.; Eslava, D.L.M.; Gavilán, P.R.; Ríos, P.M. Cambios en las actividades instrumentales de la vida diaria en la Enfermedad de Alzheimer. Acta Neurol. Colomb. 2010, 26, 112–121. [Google Scholar]
- Jefferson, A.L.; Byerly, L.K.; Vanderhill, S.; Lambe, S.; Wong, S.; Ozonoff, A.; Karlawish, J.H. Characterization of activities of daily living in individuals with mild cognitive impairment. Am. J. Geriatr. Psychiatry 2008, 16, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Gil, L.; Ruiz de Sánchez, C.; Gil, F.; Romero, S.J.; Pretelt Burgos, F. Validation of the Montreal Cognitive Assessment (MoCA) in Spanish as a screening tool for mild cognitive impairment and mild dementia in patients over 65 years old in Bogotá, Colombia. Int. J. Geriatr. Psychiatry 2015, 30, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Romero-Vanegas, S.; Romero-Vanegas, S.; Vargas-Gonzalez, J.C.; Arboleda, H.; Lopera, F.; Pardo, R. Validation of the ineco frontal screening in a colombian population. Alzheimer’s Dement. 2014, 10, P726. [Google Scholar] [CrossRef]
- Angulo, C.B.G.; Arias, A.C. Escala de Yesavage para Depresión Geriátrica (GDS-15 y GDS-5) estudio de la consistencia interna y estructura factorial. Univ. Psychol. 2011, 10, 735–743. [Google Scholar]
- Cummings, J.L.; Mega, M.; Gray, K.; Rosenberg-Thompson, S.; Carusi, D.A.; Gornbein, J. The Neuropsychiatric Inventory: Comprehensive assessment of psychopathology in dementia. Neurology 1994, 44, 2308–2314. [Google Scholar] [CrossRef]
- Peña-Casanova, J.; Casals-Coll, M.; Quintana, M.; Sánchez-Benavides, G.; Rognoni, T.; Calvo, L.; Palomo, R.; Aranciva, F.; Tamayo, F.; Manero, R.M. Estudios normativos españoles en población adulta joven (Proyecto NEURONORMA jóvenes): Métodos y características de la muestra. Neurología 2012, 27, 253–260. [Google Scholar] [CrossRef]
- Schinka, J.A.; Loewenstein, D.A.; Raj, A.; Schoenberg, M.R.; Banko, J.L.; Potter, H.; Duara, R. Defining Mild Cognitive Impairment: Impact of Varying Decision Criteria on Neuropsychological Diagnostic Frequencies and Correlates. Am. J. Geriatr. Psychiatry 2010, 18, 684–691. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.J.; Statham, A.; Stirzaker, C.; Molloy, P.L.; Frommer, M. DNA methylation: Bisulphite modification and analysis. Nat. Protoc. 2006, 1, 2353. [Google Scholar] [CrossRef]
- Tusnady, G.E.; Simon, I.; Varadi, A.; Aranyi, T. BiSearch: Primer-design and search tool for PCR on bisulfite-treated genomes. Nucleic Acids Res. 2005, 33, e9. [Google Scholar] [CrossRef]
- Hernández, H.G.; Tse, M.Y.; Pang, S.C.; Arboleda, H.; Forero, D.A. Optimizing methodologies for PCR-based DNA methylation analysis. Biotechniques 2013, 55, 181–197. [Google Scholar] [CrossRef]
- Lewin, J.; Schmitt, A.O.; Adorján, P.; Hildmann, T.; Piepenbrock, C. Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics 2004, 20, 3005–3012. [Google Scholar] [CrossRef] [Green Version]
- Mastroeni, D.; Grover, A.; Delvaux, E.; Whiteside, C.; Coleman, P.D.; Rogers, J. Epigenetic changes in Alzheimer’s disease: Decrements in DNA methylation. Neurobiol. Aging 2010, 31, 2025–2037. [Google Scholar] [CrossRef]


| Baseline Characteristics | Whole Sample | MCI | Control | p Value * |
|---|---|---|---|---|
| (n = 100) | (n = 41) | (n = 59) | ||
| Demographic data | ||||
| Age, years | ||||
| Average | 68.9 ± 9.5 | 66.5 ± 9.6 | 70.5 ± 9.1 | 0.029 |
| Range | 43–91 | 43–91 | 50–88 | |
| Gender | 0.008 | |||
| Women, n (%) | 71 (71.0) | 35 (85.4) | 36 (61.0) | |
| Men, n (%) | 29 (29.0) | 6 (14.6) | 23 (39.0) | |
| Genetic traits | ||||
| APOE-ε4 | 25 (25.0) | 10 (24.4) | 15 (25.4) | 0.999 |
| Variables | Genomic Position (hg38) | Whole Sample | MCI | Control | p Value * |
|---|---|---|---|---|---|
| (n = 100) | (n = 41) | (n = 59) | |||
| ApoE plasmatic Levels, mcg/mL | - | 103.2 ± 26.5 | 113.8 ± 26.4 | 86.0 ± 15.7 | <0.0001 † |
| Global methylation | - | 91.9 ± 3.0 | 92.8 ± 2.6 | 91.6 ± 3.1 | 0.154 † |
| Methylation by CpG sites | |||||
| CpG118 | chr19:44,909,208 | 85.6 ± 5.1 | 89.6 ± 4.1 | 84.7 ± 4.9 | 0.009 † |
| CpG130 | chr19:44,909,220 | 88.9 ± 7.1 | 89.0 ± 3.9 | 88.9 ± 7.6 | 0.484 |
| CpG133 | chr19:44,909,223 | 87.9 ± 10.1 | 85.7 ± 13.3 | 88.4 ± 9.3 | 0.620 |
| CpG148 | chr19:44,909,238 | 94.2 ± 4.2 | 94.9 ± 3.4 | 93.9 ± 4.4 | 0.255 |
| CpG162 | chr19:44,909,252 | 89.7 ± 6.1 | 90.9 ± 3.9 | 89.3 ± 6.7 | 0.361 |
| CpG165 | chr19:44,909,254 | 92 ± 5.2 | 94.5 ± 2.3 | 91.2 ± 5.6 | 0.040 † |
| CpG182 | chr19:44,909,272 | 95.2 ± 8.2 | 93.3 ± 11.7 | 95.9 ± 6.6 | 0.324 |
| CpG190 | chr19:44,909,280 | 93.8 ± 5.8 | 96.4 ± 2.3 | 93.0 ± 6.3 | 0.045 † |
| CpG198 | chr19:44,909,288 | 90.8 ± 7.9 | 96.2 ± 3.1 | 89.3 ± 8.2 | 0.01 † |
| CpG213 | chr19:44,909,303 | 95.1 ± 6.7 | 96.6 ± 6.8 | 94.7 ± 6.7 | 0.212 |
| CpG215 | chr19:44,909,305 | 90.7 ± 11.5 | 91.3 ± 12.9 | 90.5 ± 11.2 | 0.272 |
| CpG227 | chr19:44,909,317 | 97.7 ± 2.3 | 95.6 ± 2.5 | 98.4 ± 1.8 | <0.0001 |
| CpG243 | chr19:44,909,333 | 91.9 ± 10.4 | 92.5 ± 11.9 | 91.6 ± 9.9 | 0.157 |
| CpG252 | chr19:44,909,342 | 90.4 ± 6.2 | 90.0 ± 6.1 | 90.6 ± 6.4 | 0.833 |
| Variables | Non-APOE-ε4 Carriers | APOE-ε4 Carriers | p Value * |
|---|---|---|---|
| (n = 70) | (n = 25) | ||
| ApoE plasma levels, mcg/mL | 106.0 ± 31.3 | 103.0 ± 26.2 | 0.738 † |
| Global methylation | 92.2 ± 2.2 | 91.6 ± 3.3 | 0.557 † |
| Methylation by CpG Sites | |||
| CpG118 | 84.5 ± 6.0 | 85.8 ± 4.5 | 0.428 † |
| CpG130 | 90.9 ± 6.5 | 88.0 ± 7.3 | 0.079 |
| CpG133 | 84.8 ± 11.9 | 89.9 ± 7.0 | 0.087 |
| CpG148 | 96.2 ± 2.9 | 93.3 ± 4.4 | 0.003 |
| CpG162 | 90.8 ± 3.1 | 89.4 ± 7.0 | 0.447 |
| CpG165 | 89.7 ± 4.3 | 92.6 ± 5.4 | 0.076 † |
| CpG182 | 94.2 ± 10.1 | 95.4 ± 7.7 | 0.474 |
| CpG190 | 95.0 ± 2.0 | 94.1 ± 3.1 | 0.316 † |
| CpG198 | 91.1 ± 4.6 | 91.4 ± 5.6 | 0.860 † |
| CpG213 | 95.6 ± 5.2 | 94.8 ± 7.4 | 0.872 |
| CpG215 | 93.0 ± 8.1 | 89.6 ± 12.7 | 0.650 |
| CpG227 | 97.5 ± 2.4 | 98.0 ± 2.1 | 0.482 |
| CpG243 | 95.0 ± 3.6 | 90.0 ± 12.3 | 0.363 |
| CpG252 | 89.9 ± 3.5 | 90.5 ± 7.3 | 0.200 |
| Variables | Risk for Mild Cognitive Impairment | ||
|---|---|---|---|
| Odds Ratios | 95% Confident Interval | p Value | |
| Plasma ApoE levels | 1.07 | 1.02–1.13 | 0.003 |
| APOE methylation | - | ||
| CpG 118 | 1.25 | 0.96–1.62 | 0.092 |
| CpG 165 | 1.20 | 1.01–1.43 | 0.045 |
| CpG 190 | 1.52 | 1.06–2.19 | 0.023 |
| CpG 198 | 1.30 | 1.01–1.67 | 0.042 |
| CpG 227 * | 10.05 | 1.50–67.30 | 0.017 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mancera-Páez, O.; Estrada-Orozco, K.; Mahecha, M.F.; Cruz, F.; Bonilla-Vargas, K.; Sandoval, N.; Guerrero, E.; Salcedo-Tacuma, D.; Melgarejo, J.D.; Vega, E.; et al. Differential Methylation in APOE (Chr19; Exon Four; from 44,909,188 to 44,909,373/hg38) and Increased Apolipoprotein E Plasma Levels in Subjects with Mild Cognitive Impairment. Int. J. Mol. Sci. 2019, 20, 1394. https://doi.org/10.3390/ijms20061394
Mancera-Páez O, Estrada-Orozco K, Mahecha MF, Cruz F, Bonilla-Vargas K, Sandoval N, Guerrero E, Salcedo-Tacuma D, Melgarejo JD, Vega E, et al. Differential Methylation in APOE (Chr19; Exon Four; from 44,909,188 to 44,909,373/hg38) and Increased Apolipoprotein E Plasma Levels in Subjects with Mild Cognitive Impairment. International Journal of Molecular Sciences. 2019; 20(6):1394. https://doi.org/10.3390/ijms20061394
Chicago/Turabian StyleMancera-Páez, Oscar, Kelly Estrada-Orozco, María Fernanda Mahecha, Francy Cruz, Kely Bonilla-Vargas, Nicolás Sandoval, Esneyder Guerrero, David Salcedo-Tacuma, Jesús D. Melgarejo, Edwin Vega, and et al. 2019. "Differential Methylation in APOE (Chr19; Exon Four; from 44,909,188 to 44,909,373/hg38) and Increased Apolipoprotein E Plasma Levels in Subjects with Mild Cognitive Impairment" International Journal of Molecular Sciences 20, no. 6: 1394. https://doi.org/10.3390/ijms20061394