Gymnotic Delivery of LNA Mixmers Targeting Viral SREs Induces HIV-1 mRNA Degradation
Abstract
:1. Introduction
2. Results
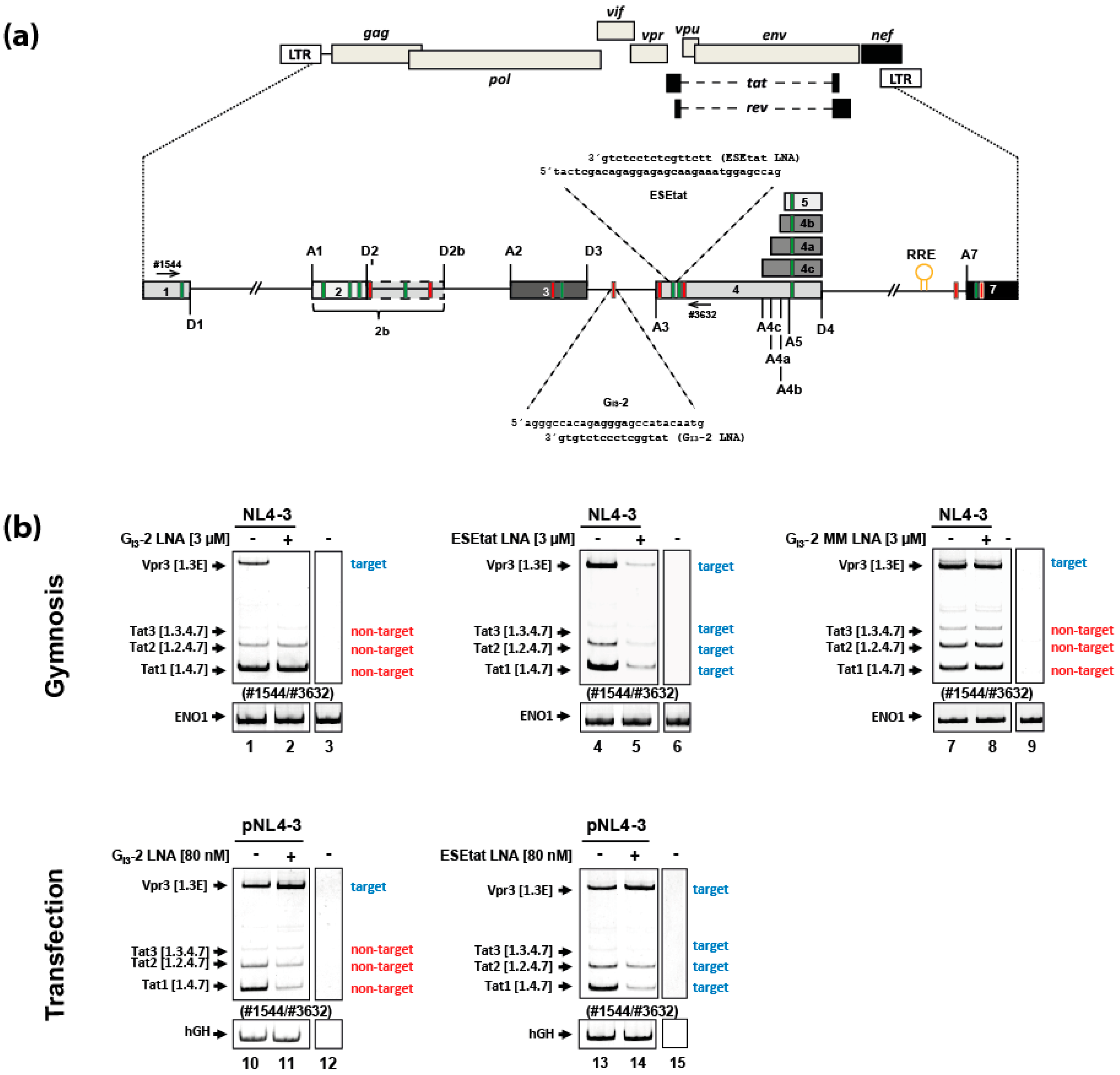
2.1. Gymnotically-Delivered LNA Mixmers Binding the SREs GI3-2 and ESEtat Specifically Induce Degradation of Their Target mRNAs
2.2. A Gymnotically-Delivered SRSF6 Exon/Junction LNA Mixmer Induces Degradation of the SRSF6 mRNA
2.3. The FAM-Labeled SRSF6 ExJ 3/4 LNA Mixmer Co-Localizes with GW-182 in HeLa and Jurkat Cells
2.4. Gymnotic Delivery of both LNA Mixmers, GI3-2 and ESEtat, Efficiently Interferes with Viral RNA Expression and HIV-1 Replication in Infected T-Cells
2.5. The Antiretroviral Effect of the GI3-2 and ESEtat LNA Mixmers as Well as a Cocktail Consisting of Both LNAs Lasts up to Nearly 14 Days in Jurkat Cells and in PBMCs
3. Discussion
4. Materials and Methods
4.1. Oligonucleotides
4.2. Cell Culture, Preparation of Virus Stocks, Infection Experiments and Gymnotic LNA Delivery
4.3. Transfection of LNA Antisense Oligonucleotides
4.4. RNA and Protein Isolation
4.5. Immunoblot Analysis
4.6. RT-PCR-Analysis
4.7. Northern Blot Analysis
4.8. Confocal Laser Scanning Microscopy
4.8.1. Intracellular Localization of 6-FAM™-Labeled LNAs
4.8.2. Co-Localization of Gymnotically-Delivered 6-FAM™-Labeled LNAs with GW-182
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2′-MOE | 2′-O-methoxyethyl |
| 2′-OMe | 2′-O-methyl |
| 6-FAM™ | 6-Carboxyfluorescein |
| Ago-2 | protein argonaute-2 |
| ApoB | Apolipoprotein B |
| ART | antiretroviral therapy |
| ASO | antisense oligonucleotide |
| Bcl-2 | Apoptosis regulator Bcl-2 |
| CA | capsid |
| CCR4–NOT | carbon catabolite repressor 4- negative on TATA |
| D3 | splice donor 3 |
| DCP1/2 | mRNA-decapping enzyme 1/2 |
| DMD | Duchenne muscular dystrophy |
| ERK2 | mitogen-activated protein kinase 1 |
| ESEtat | exonic splicing enhancer tat |
| ExJ 3/4 | exon junction exon3/exon4 |
| FDA | U.S. Food and Drug Administration |
| Gapmer | LNAs at the 5′- and 3′ end and a DNA strand in the center of the ASO |
| GAR | guanosine-adenosine-rich exonic splicing enhancer |
| GI3-2 | second G-run within HIV-1 intron 3 |
| GW-182 | glycine-tryptophan protein of 182 kDa |
| GW-body | cytoplasmic foci containing enzymes involved in RNA degradation and translational repression |
| HCV | hepatitis C virus |
| HIV-1 | human immunodeficiency virus type 1 |
| hnRNP | Heterogeneous nuclear ribonucleoproteins |
| IL-2 | Interleukin-2 |
| LNA | Locked nucleic acid |
| miRNA | microRNA |
| Mixmer | mixed combination of LNA and DNA residues within the ASO |
| PAZ domain | Piwi/Argonaute/Zwille domain |
| PBMC | peripheral blood mononuclear cell |
| PMO | phosphorodiamidate morpholinos |
| PS | phosphorothioate |
| RNAi | RNA interference |
| RRE | Rev responsive element |
| SA3 | splice acceptor 3 |
| SIRC | stress-induced response complex |
| siRNA | small interfering RNA |
| SMA | spinal muscular atrophy |
| SR | serine and arginine-rich protein |
| SRE | splicing regulatory element |
| SRSF6 | serine/arginine-rich splicing factor 6 |
| SSO | splice-switching oligonucleotide |
| TAR | trans-activation response element |
| U1 snRNA | U1 small nuclear ribonucleic acid |
| WHO | World Health Organization |
| XRN1 | 5’-3’ exoribonuclease 1 |
References
- Pennings, P.S. HIV Drug Resistance: Problems and Perspectives. Infect. Dis. Rep. 2013, 5 (Suppl. 1), e5. [Google Scholar] [CrossRef]
- Loucif, H.; Gouard, S.; Dagenais-Lussier, X.; Murira, A.; Stager, S.; Tremblay, C.; Van Grevenynghe, J. Deciphering natural control of HIV-1: A valuable strategy to achieve antiretroviral therapy termination. Cytokine Growth Factor Rev. 2018, 40, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Purcell, D.F.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [PubMed]
- Widera, M.; Erkelenz, S.; Hillebrand, F.; Krikoni, A.; Widera, D.; Kaisers, W.; Deenen, R.; Gombert, M.; Dellen, R.; Pfeiffer, T.; et al. An intronic G-run within HIV-1 intron 2 is critical for splicing regulation of vif-mRNA. J. Virol. 2013, 87, 2707–2720. [Google Scholar] [CrossRef] [PubMed]
- Stoltzfus, C.M. Chapter 1. Regulation of HIV-1 alternative RNA splicing and its role in virus replication. Adv. Virus Res. 2009, 74, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Ocwieja, K.E.; Sherrill-Mix, S.; Mukherjee, R.; Custers-Allen, R.; David, P.; Brown, M.; Wang, S.; Link, D.R.; Olson, J.; Travers, K.; et al. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res. 2012, 40, 10345–10355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emery, A.; Zhou, S.; Pollom, E.; Swanstrom, R. Characterizing HIV-1 Splicing by Using Next-Generation Sequencing. J. Virol. 2017, 91, e02515-16. [Google Scholar] [CrossRef] [PubMed]
- Sertznig, H.; Hillebrand, F.; Erkelenz, S.; Schaal, H.; Widera, M. Behind the scenes of HIV-1 replication: Alternative splicing as the dependency factor on the quiet. Virology 2018, 516, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Erkelenz, S.; Mueller, W.F.; Evans, M.S.; Busch, A.; Schoneweis, K.; Hertel, K.J.; Schaal, H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 2012, 19, 96–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madsen, J.M.; Stoltzfus, C.M. An exonic splicing silencer downstream of the 3’ splice site A2 is required for efficient human immunodeficiency virus type 1 replication. J. Virol. 2005, 79, 10478–10486. [Google Scholar] [CrossRef] [PubMed]
- Exline, C.M.; Feng, Z.; Stoltzfus, C.M. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J. Virol. 2008, 82, 3921–3931. [Google Scholar] [CrossRef] [PubMed]
- Asang, C.; Hauber, I.; Schaal, H. Insights into the selective activation of alternatively used splice acceptors by the human immunodeficiency virus type-1 bidirectional splicing enhancer. Nucleic Acids Res. 2008, 36, 1450–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widera, M.; Hillebrand, F.; Erkelenz, S.; Vasudevan, A.; Munk, C.; Schaal, H. A functional conserved intronic G run in HIV-1 intron 3 is critical to counteract APOBEC3G-mediated host restriction. Retrovirology 2014, 11, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erkelenz, S.; Poschmann, G.; Theiss, S.; Stefanski, A.; Hillebrand, F.; Otte, M.; Stuhler, K.; Schaal, H. Tra2-mediated recognition of HIV-1 5’ss D3 as a key factor in processing vpr-mRNA. J. Virol. 2013, 87, 721–2734. [Google Scholar] [CrossRef] [PubMed]
- Erkelenz, S.; Hillebrand, F.; Widera, M.; Theiss, S.; Fayyaz, A.; Degrandi, D.; Pfeffer, K.; Schaal, H. Balanced splicing at the Tat-specific HIV-1 3’ss A3 is critical for HIV-1 replication. Retrovirology 2015, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Brillen, A.L.; Walotka, L.; Hillebrand, F.; Muller, L.; Widera, M.; Theiss, S.; Schaal, H. Analysis of competing HIV-1 splice donor sites uncovers a tight cluster of splicing regulatory elements within exon 2/2b. J. Virol. 2017, 91, e00389-17. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, J.; Agrawal, S.; Civeira, M.P.; Sarin, P.S.; Sun, D.; Zamecnik, P.C. Inhibition of human immunodeficiency virus replication by antisense oligodeoxynucleotides. Proc. Natl. Acad. Sci. USA 1988, 85, 5507–5511. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, M.; Zon, G.; Shinozuka, K.; Robert-Guroff, M.; Shimada, T.; Stein, C.A.; Mitsuya, H.; Wong-Staal, F.; Cohen, J.S.; Broder, S. Regulation of viral expression of human immunodeficiency virus in vitro by an antisense phosphorothioate oligodeoxynucleotide against rev (art/trs) in chronically infected cells. Proc. Natl. Acad. Sci. USA 1989, 86, 4244–4248. [Google Scholar] [CrossRef] [PubMed]
- Arzumanov, A.; Walsh, A.P.; Rajwanshi, V.K.; Kumar, R.; Wengel, J.; Gait, M.J. Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2’-O-methyl/LNA oligoribonucleotides. Biochemistry 2001, 40, 14645–14654. [Google Scholar] [CrossRef] [PubMed]
- Lebars, I.; Richard, T.; Di Primo, C.; Toulme, J.J. LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood Cells Mol. Dis. 2007, 38, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.R.; Haasnoot, J.; Wengel, J.; Berkhout, B.; Kjems, J. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology 2007, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asparuhova, M.B.; Marti, G.; Liu, S.; Serhan, F.; Trono, D.; Schumperli, D. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J. Gene Med. 2007, 9, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Havens, M.A.; Hastings, M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooke, S.T.; Witztum, J.L.; Bennett, C.F.; Baker, B.F. RNA-Targeted Therapeutics. Cell Metab. 2018, 27, 714–739. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Vickers, T.A.; Okunola, H.L.; Bennett, C.F.; Krainer, A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008, 82, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.N.; Howell, M.D.; Androphy, E.J.; Singh, R.N. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017, 24, 520–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunweller, A.; Hartmann, R.K. Locked nucleic acid oligonucleotides: The next generation of antisense agents? Biodrugs Clin. Immunother. Biopharm. Gene Ther. 2007, 21, 235–243. [Google Scholar] [CrossRef]
- Lundin, K.E.; Hojland, T.; Hansen, B.R.; Persson, R.; Bramsen, J.B.; Kjems, J.; Koch, T.; Wengel, J.; Smith, C.I. Biological activity and biotechnological aspects of locked nucleic acids. Adv. Genet. 2013, 82, 47–107. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A.; Hansen, J.B.; Lai, J.; Wu, S.; Voskresenskiy, A.; Hog, A.; Worm, J.; Hedtjarn, M.; Souleimanian, N.; Miller, P.; et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010, 38, e3. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qu, Z.; Kim, S.; Shi, V.; Liao, B.; Kraft, P.; Bandaru, R.; Wu, Y.; Greenberger, L.M.; Horak, I.D. Down-modulation of cancer targets using locked nucleic acid (LNA)-based antisense oligonucleotides without transfection. Gene Ther. 2011, 18, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Fazil, M.H.; Ong, S.T.; Chalasani, M.L.; Low, J.H.; Kizhakeyil, A.; Mamidi, A.; Lim, C.F.; Wright, G.D.; Lakshminarayanan, R.; Kelleher, D.; et al. GapmeR cellular internalization by macropinocytosis induces sequence-specific gene silencing in human primary T-cells. Sci. Rep. 2016, 6, 37721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souleimanian, N.; Deleavey, G.F.; Soifer, H.; Wang, S.; Tiemann, K.; Damha, M.J.; Stein, C.A. Antisense 2’-Deoxy, 2’-Fluroarabino Nucleic Acids (2’F-ANAs) Oligonucleotides: In Vitro Gymnotic Silencers of Gene Expression Whose Potency Is Enhanced by Fatty Acids. Mol. Ther. Nucleic Acids 2012, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- Castanotto, D.; Lin, M.; Kowolik, C.; Wang, L.; Ren, X.Q.; Soifer, H.S.; Koch, T.; Hansen, B.R.; Oerum, H.; Armstrong, B.; et al. A cytoplasmic pathway for gapmer antisense oligonucleotide-mediated gene silencing in mammalian cells. Nucleic Acids Res. 2015, 43, 9350–9361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akhtar, S.; Juliano, R.L. Cellular uptake and intracellular fate of antisense oligonucleotides. Trends Cell Biol. 1992, 2, 139–144. [Google Scholar] [CrossRef]
- Koller, E.; Vincent, T.M.; Chappell, A.; De, S.; Manoharan, M.; Bennett, C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011, 39, 4795–4807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juliano, R.L. Intracellular Trafficking and Endosomal Release of Oligonucleotides: What We Know and What We Don’t. Nucleic Acid Ther. 2018, 28, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Ottosen, S.; Parsley, T.B.; Yang, L.; Zeh, K.; van Doorn, L.J.; van der Veer, E.; Raney, A.K.; Hodges, M.R.; Patick, A.K. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob. Agents Chemother. 2015, 59, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Morelli, E.; Biamonte, L.; Federico, C.; Amodio, N.; Di Martino, M.T.; Gallo Cantafio, M.E.; Manzoni, M.; Scionti, F.; Samur, M.K.; Gulla, A.; et al. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-mir-17-92. Blood 2018, 132, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Shen, W.; Sun, H.; Prakash, T.P.; Crooke, S.T. TCP1 complex proteins interact with phosphorothioate oligonucleotides and can co-localize in oligonucleotide-induced nuclear bodies in mammalian cells. Nucleic Acids Res. 2014, 42, 7819–7832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, J.K.; Shen, W.; Liang, X.H.; Crooke, S.T. Nucleic acid binding proteins affect the subcellular distribution of phosphorothioate antisense oligonucleotides. Nucleic Acids Res. 2017, 45, 10649–10671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.H.; Sun, H.; Shen, W.; Crooke, S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015, 43, 2927–2945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, T.A.; Crooke, S.T. Development of a Quantitative BRET Affinity Assay for Nucleic Acid-Protein Interactions. PLoS ONE 2016, 11, e0161930. [Google Scholar] [CrossRef] [PubMed]
- Castanotto, D.; Zhang, X.; Alluin, J.; Zhang, X.; Ruger, J.; Armstrong, B.; Rossi, J.; Riggs, A.; Stein, C.A. A stress-induced response complex (SIRC) shuttles miRNAs, siRNAs, and oligonucleotides to the nucleus. Proc. Natl. Acad. Sci. USA 2018, 115, E5756–E5765. [Google Scholar] [CrossRef] [PubMed]
- Kurreck, J.; Wyszko, E.; Gillen, C.; Erdmann, V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frieden, M.; Christensen, S.M.; Mikkelsen, N.D.; Rosenbohm, C.; Thrue, C.A.; Westergaard, M.; Hansen, H.F.; Orum, H.; Koch, T. Expanding the design horizon of antisense oligonucleotides with alpha-L-LNA. Nucleic Acids Res. 2003, 31, 6365–6372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakymiw, A.; Lian, S.; Eystathioy, T.; Li, S.; Satoh, M.; Hamel, J.C.; Fritzler, M.J.; Chan, E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005, 7, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Siomi, M.C. RISC hitches onto endosome trafficking. Nat. Cell Biol. 2009, 11, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Belasco, J.G. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell 2008, 29, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]




| LNA Oligonucleotide | Sequence | Design-ID (Exiqon) | Cat. No Qiagen |
|---|---|---|---|
| GI3-2 [13] | TATGGCTCCCTCTGTG | 164610 | YCO0073444 |
| ESEtat [15] | TTCTTGCTCTCCTCTG | 256589 | YCO0073445 |
| SRSF6 D3 (5’-end modiefied with 6-FAM™) | TACAAAACATACCTTT | 319384 | - |
| SRSF6 ExJ 3/4 (5’-end modiefied with 6-FAM™) | TCGCATAAAATCCTTT | 548164 | - |
| GI3-1-MM-control | TTTGGCTCACTCCGTG | 240758 | - |
| mRNA Type | Primer No. | Primer Sequence |
|---|---|---|
| HIV-1 exon1-4 mRNAs | #1544 (exon1) | 5’ CTTGAAAGCGAAAGTAAAGC 3’ |
| #3632 (exon4) | 5’ TGGATGCTTCCAGGGCTC 3’ | |
| HIV-1 exon7 | #3387 | 5’ TTGCTCAATG CCACAGCCAT 3’ |
| #3388 | 5’ TTTGACCACT TGCCACCCAT 3’ | |
| SRSF6 mRNA | #4933 | 5’ GAGTTCGAGGACTCCCG 3’ |
| #4934 | 5’ TCTACTGCGGCTGCTCCT 3’ | |
| ENO1 mRNA | #4907 | 5’ CTGTGCCCAGTGGTGCT 3’ |
| #4908 | 5’ GACCTGAAGAACTCGGAGG 3’ | |
| hGH mRNA | #1224 | 5’ TCTTCCAGCCTCCCATCAGCGTTTGG 3’ |
| #1225 | 5’ CAACAGAAATCCAACCTAGAGCTGCT 3’ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hillebrand, F.; Ostermann, P.N.; Müller, L.; Degrandi, D.; Erkelenz, S.; Widera, M.; Pfeffer, K.; Schaal, H. Gymnotic Delivery of LNA Mixmers Targeting Viral SREs Induces HIV-1 mRNA Degradation. Int. J. Mol. Sci. 2019, 20, 1088. https://doi.org/10.3390/ijms20051088
Hillebrand F, Ostermann PN, Müller L, Degrandi D, Erkelenz S, Widera M, Pfeffer K, Schaal H. Gymnotic Delivery of LNA Mixmers Targeting Viral SREs Induces HIV-1 mRNA Degradation. International Journal of Molecular Sciences. 2019; 20(5):1088. https://doi.org/10.3390/ijms20051088
Chicago/Turabian StyleHillebrand, Frank, Philipp Niklas Ostermann, Lisa Müller, Daniel Degrandi, Steffen Erkelenz, Marek Widera, Klaus Pfeffer, and Heiner Schaal. 2019. "Gymnotic Delivery of LNA Mixmers Targeting Viral SREs Induces HIV-1 mRNA Degradation" International Journal of Molecular Sciences 20, no. 5: 1088. https://doi.org/10.3390/ijms20051088





