Description and Characterization of a Novel Human Mast Cell Line for Scientific Study
Abstract
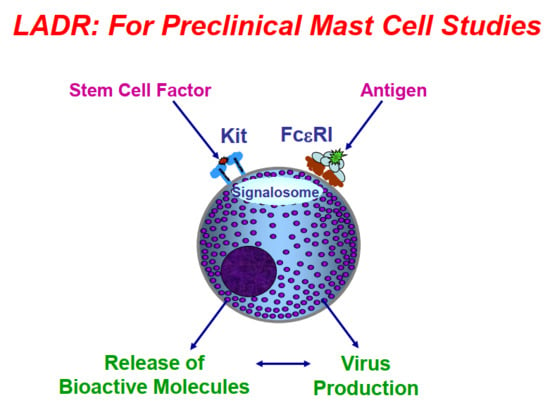
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. LAD Cell Cultures
4.2. β-Hex Release Studies
4.3. Flow Cytometry Studies
4.4. Gene Arrays
4.5. HIV Infectivity
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Butterfield, J.H.; Weiler, D.; Dewald, G.; Gleich, G.J. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leukemia Res. 1988, 12, 345–355. [Google Scholar] [CrossRef]
- Kirshenbaum, A.S.; Akin, C.; Wu, Y.; Rottem, M.; Goff, J.P.; Beaven, M.A.; Rao, K.; Metcalfe, D.D. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FceRI or FceRI. Leukemia Res. 2003, 27, 677–682. [Google Scholar] [CrossRef]
- Laidlaw, T.M.; Steinke, J.W.; Tiñana, A.M.; Feng, C.; Xing, W.; Lam, B.K.; Paruchuri, S.; Boyce, J.A.; Borish, L. Characterization of a novel human mast cell line that responds to stem cell factor and expresses functional FcεRI. J. Allergy Clin. Immun. 2011, 127, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Saleh, R.; Wedeh, G.; Herrmann, H.; Bibi, S.; Cerny-Reiterer, S.; Sadovnik, I.; Blatt, K.; Hadzijusufovic, E.; Jeanningros, S.; Blanc, C.; et al. A new human mast cell line expressing a functional IgE receptor converts to tumorigenic growth by KIT D816V transfection. Blood 2014, 124, 111–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arock, M.; Wedeh, G.; Hoermann, G.; Bibi, S.; Akin, C.; Peter, B.; Gleixner, K.V.; Hartmann, K.; Butterfield, J.H.; Metcalfe, D.D.; et al. Preclinical human models and emerging therapeutics for advanced systemic mastocytosis. Haematologica 2018, 103, 1760–1771. [Google Scholar] [CrossRef] [PubMed]
- Sundstrom, J.B.; Hair, G.A.; Ansari, A.A.; Secor, W.E.; Gilfillan, A.M.; Metcalfe, D.D.; Kirshenbaum, A.S. IgE-FcepsilonRI interactions determine HIV coreceptor usage and susceptibility to infection during ontogeny of mast cells. J. Immunol. 2009, 182, 6401–6409. [Google Scholar] [CrossRef] [PubMed]
- Radinger, M.; Jensen, B.M.; Kuehn, H.S.; Kirshenbaum, A.S.; Gilfillan, A.M. Generation, isolation, and maintenance of human mast cells and mast cell lines derived from peripheral blood or cord blood. Curr. Protoc. Immunol. 2010, 90, 7.37.1–7.37.12. [Google Scholar]
- Kuehn, H.S.; Radinger, M.; Gilfillan, A.M. Measuring mast cell mediator release. Curr. Protoc. Immunol. 2010, 91, 7.38.1–7.38.9. [Google Scholar]
- Tomonobu, I.; Smrz, D.; Jung, M.-Y.; Bandara, G.; Desai, A.; Smrzová, S.; Kuehn, H.S.; Beaven, M.A.; Metcalfe, D.D.; Gilfillan, A.M. Stem cell factor programs the mast cell activation phenotype. J. Immunol. 2012, 188, 5428–5437. [Google Scholar]
- Eunice, C.C.; Bai, Y.; Bandara, G.; Simakovab, O.; Brittain, E.; Scott, L.; Dyer, K.D.; Klion, A.D.; Maric, I.; Gilfillan, A.M.; et al. KIT GNNK splice variants: Expression in systemic mastocytosis and influence on the activating potential of the D816V mutation in mast cells. Exp. Hematol. 2013, 41, 870–881. [Google Scholar]
- Sundstrom, J.B.; Ellis, J.E.; Hair, G.A.; Kirshenbaum, A.S.; Metcalfe, D.D.; Yi, H.; Cardona, A.C.; Lindsay, M.K.; Ansari, A.A. Human tissue mast cells are an inducible reservoir of persistent HIV infection. Blood 2007, 109, 5293–5300. [Google Scholar] [CrossRef] [PubMed]





| CD Marker | LADR Cells | LAD2 Cells |
|---|---|---|
| CD13 | + + | − |
| CD25 | − | − |
| CD33 | + | + |
| CD34 | + | + |
| CD63 | + + | + + |
| CD117 | + + + | + + + |
| CD123 | + | − |
| CD133 | + | + |
| CD184 | + + | − |
| CD193 | + | + |
| CD195 | + | − |
| FcεRI | + + | + + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
S. Kirshenbaum, A.; Yin, Y.; Sundstrom, J.B.; Bandara, G.; D. Metcalfe, D. Description and Characterization of a Novel Human Mast Cell Line for Scientific Study. Int. J. Mol. Sci. 2019, 20, 5520. https://doi.org/10.3390/ijms20225520
S. Kirshenbaum A, Yin Y, Sundstrom JB, Bandara G, D. Metcalfe D. Description and Characterization of a Novel Human Mast Cell Line for Scientific Study. International Journal of Molecular Sciences. 2019; 20(22):5520. https://doi.org/10.3390/ijms20225520
Chicago/Turabian StyleS. Kirshenbaum, Arnold, Yuzhi Yin, J. Bruce Sundstrom, Geethani Bandara, and Dean D. Metcalfe. 2019. "Description and Characterization of a Novel Human Mast Cell Line for Scientific Study" International Journal of Molecular Sciences 20, no. 22: 5520. https://doi.org/10.3390/ijms20225520