Genome-Wide Analysis of Cotton miRNAs During Whitefly Infestation Offers New Insights into Plant-Herbivore Interaction
Abstract
:1. Introduction
2. Results
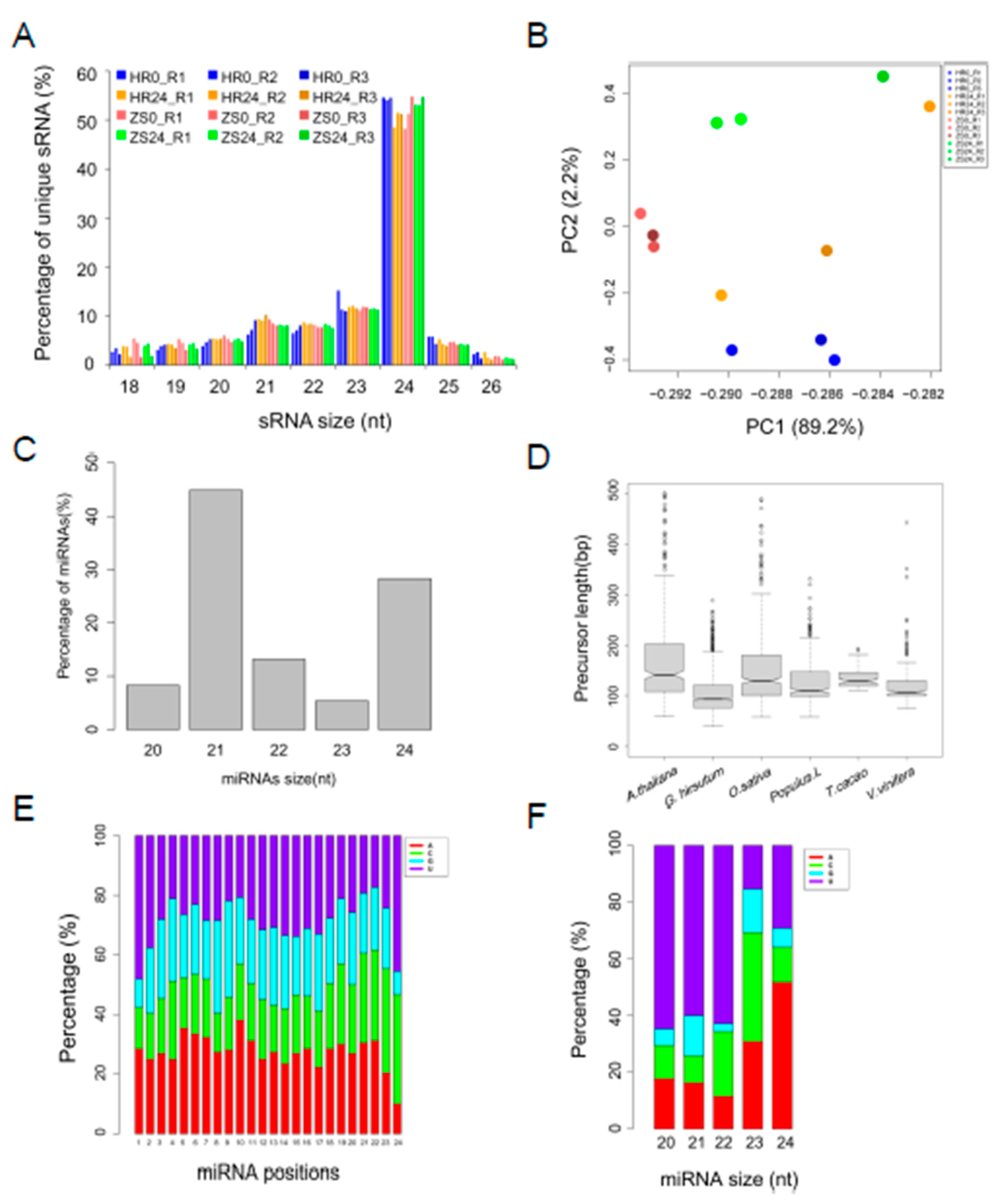
2.1. Classification and Annotation of sRNAs in Resistant (HR) and Susceptible (ZS) Cotton Cultivar in Response to Whitefly Infestation
2.2. Abundant lincRNA Act as miRNA Precursor
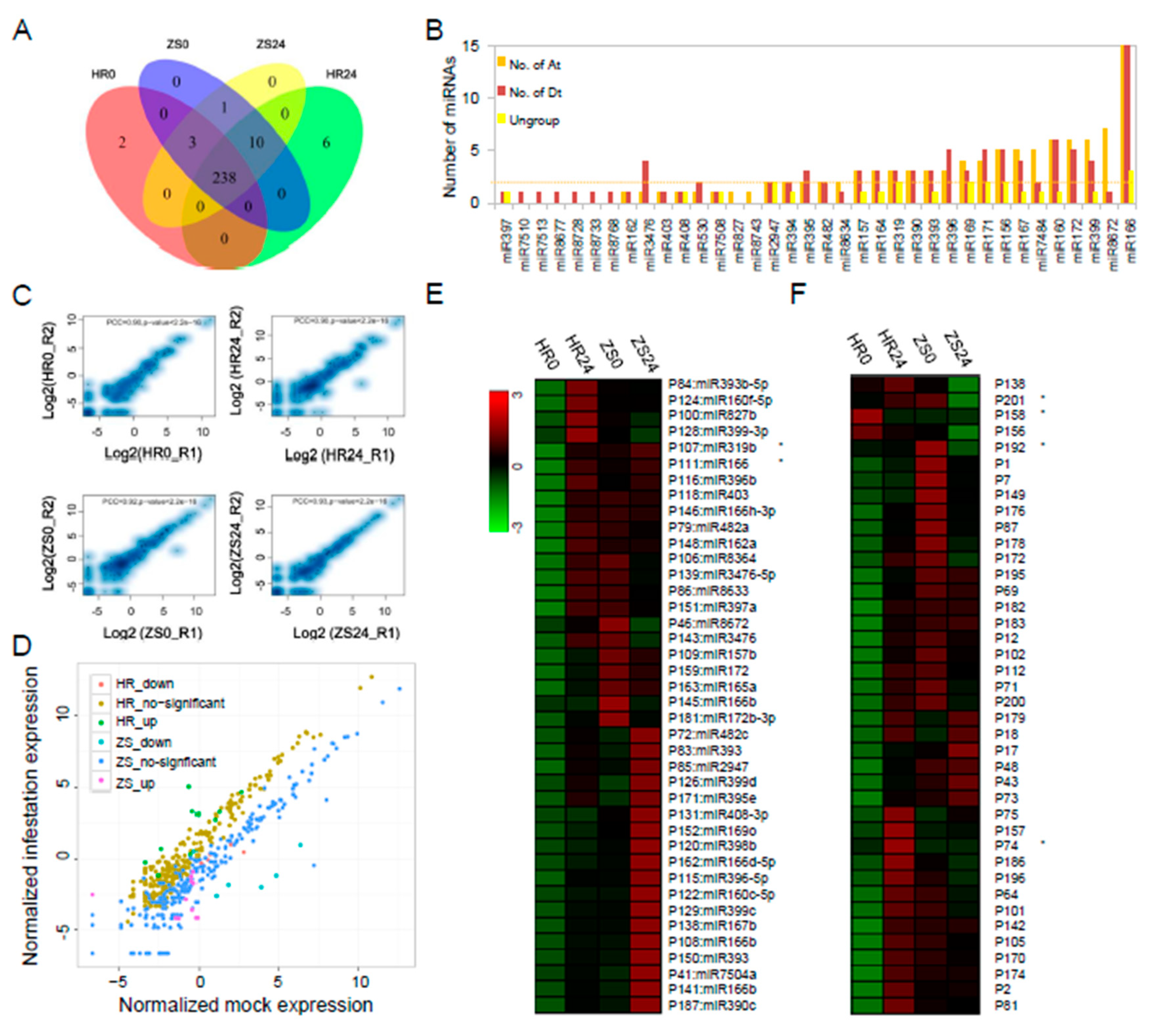
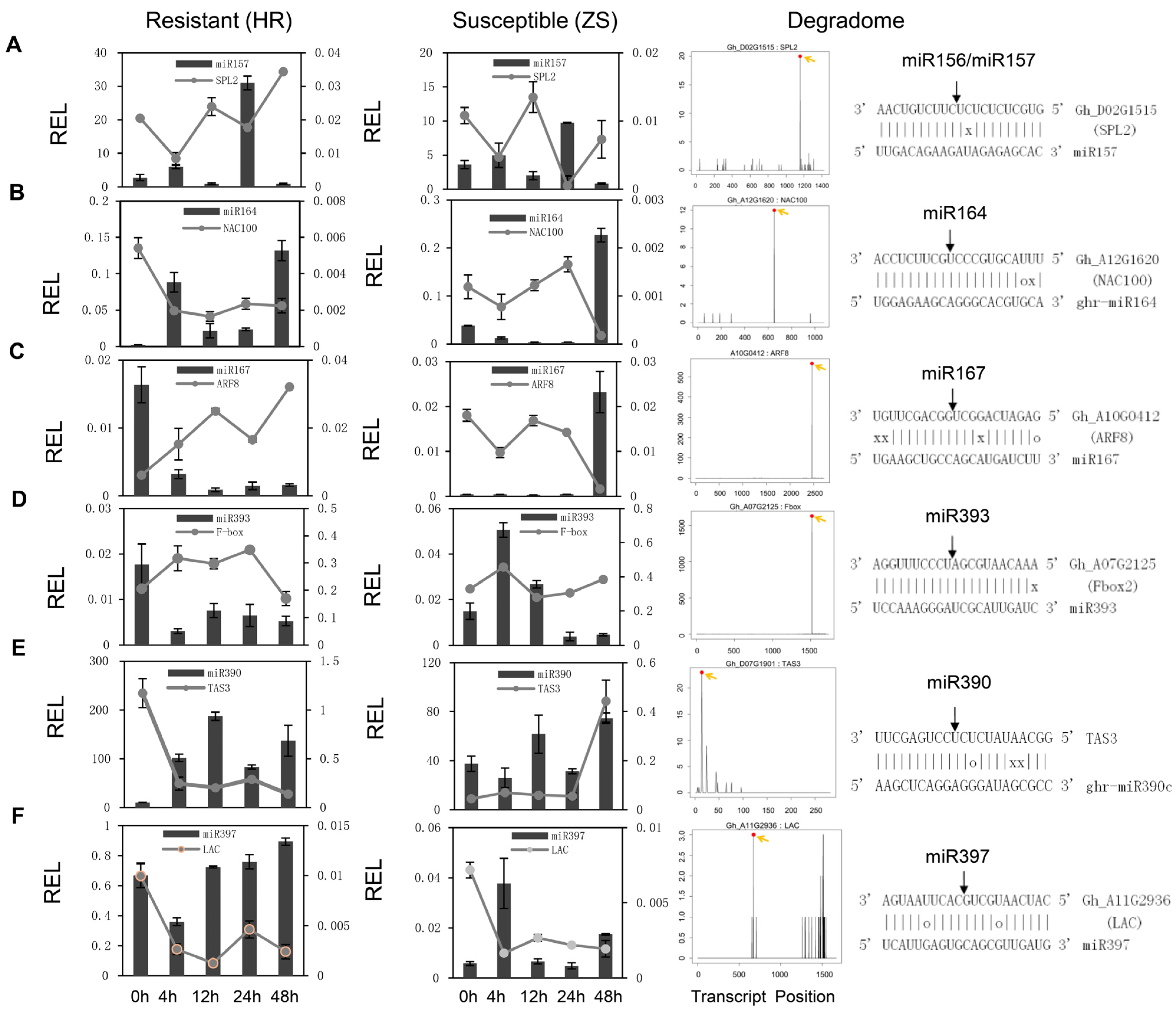
2.3. Analysis of the miRNA Target Genes Based on Degradome Sequencing
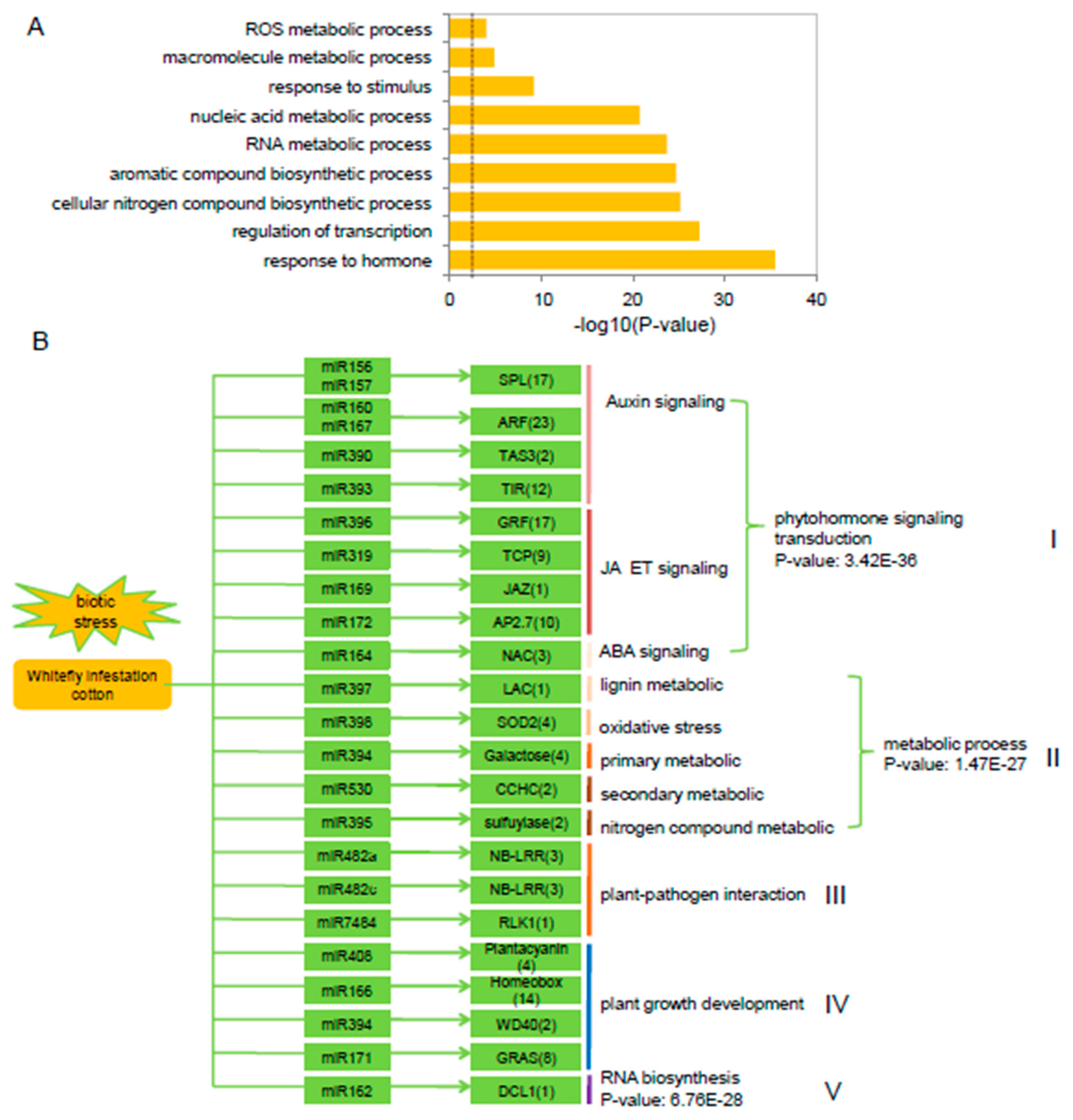
2.4. Tight Linkage Between miRNAs and Their Targets in Cotton in Response to Whitefly Infestation
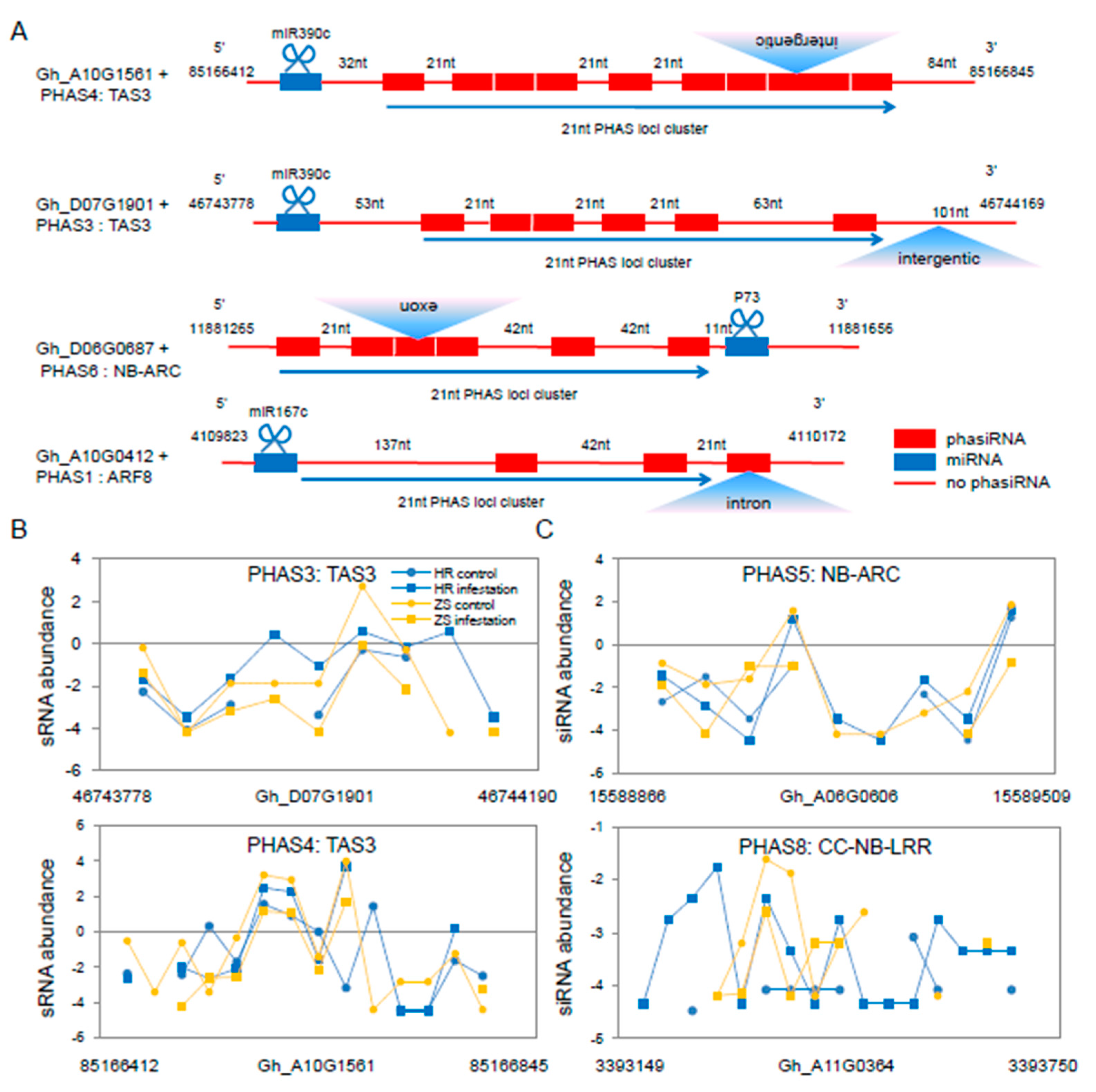
2.5. Identification of miRNA-mediated phasiRNAs during the Whitefly Infestation Cotton Plants
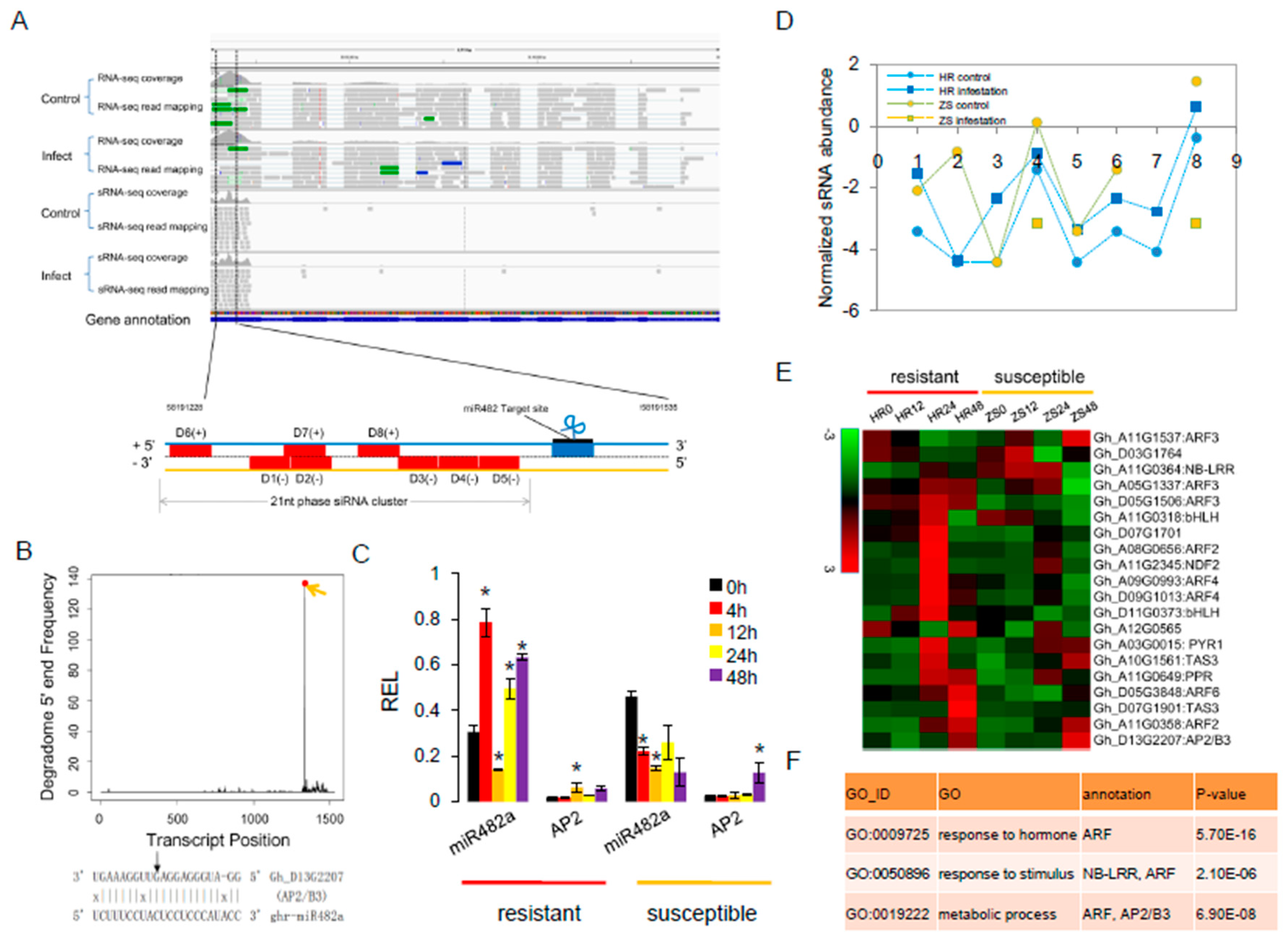
2.6. mi482a-Triggered phasiRNAs Regulate the Transcriptional in Cotton Response to Whitefly Infestation
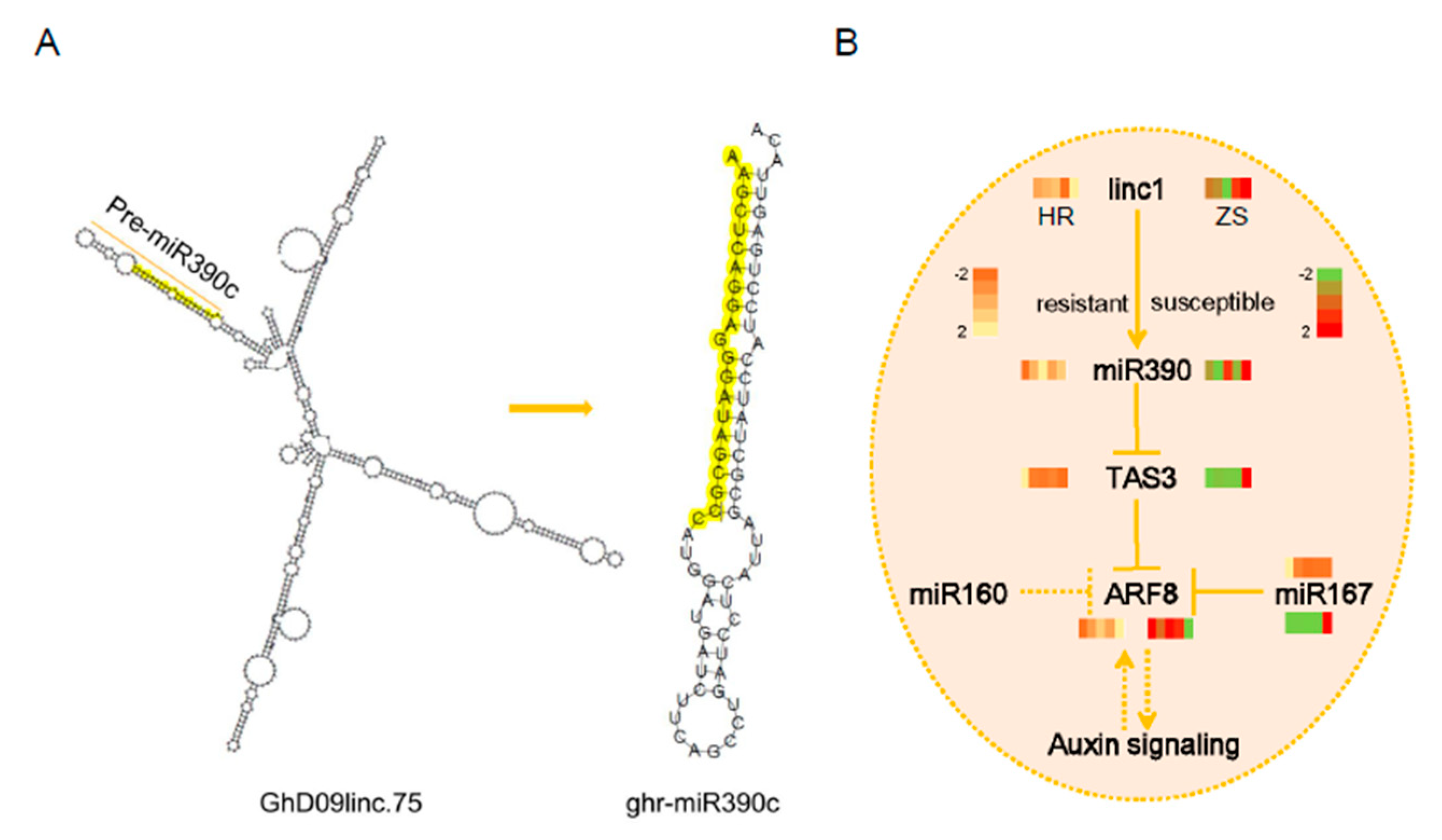
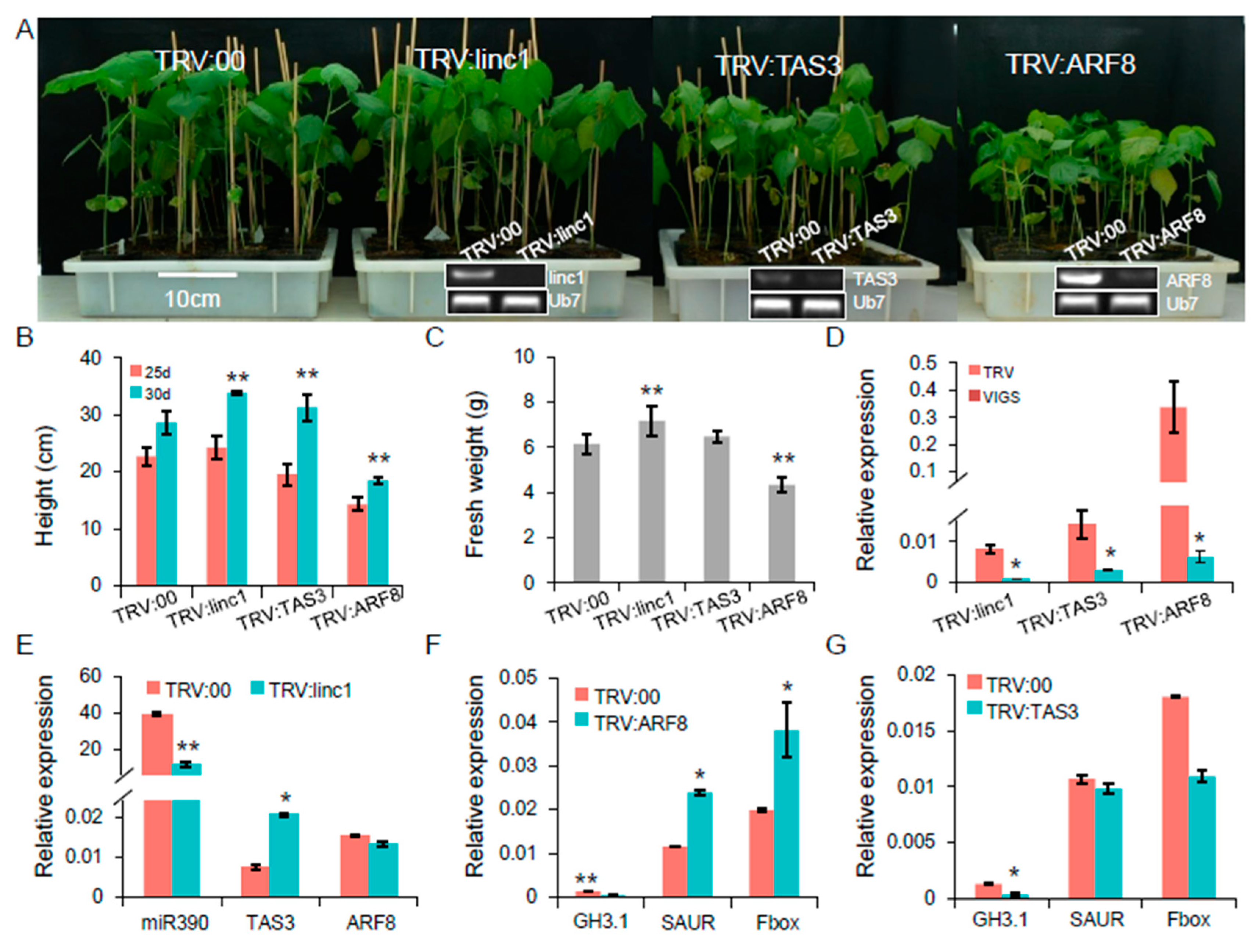
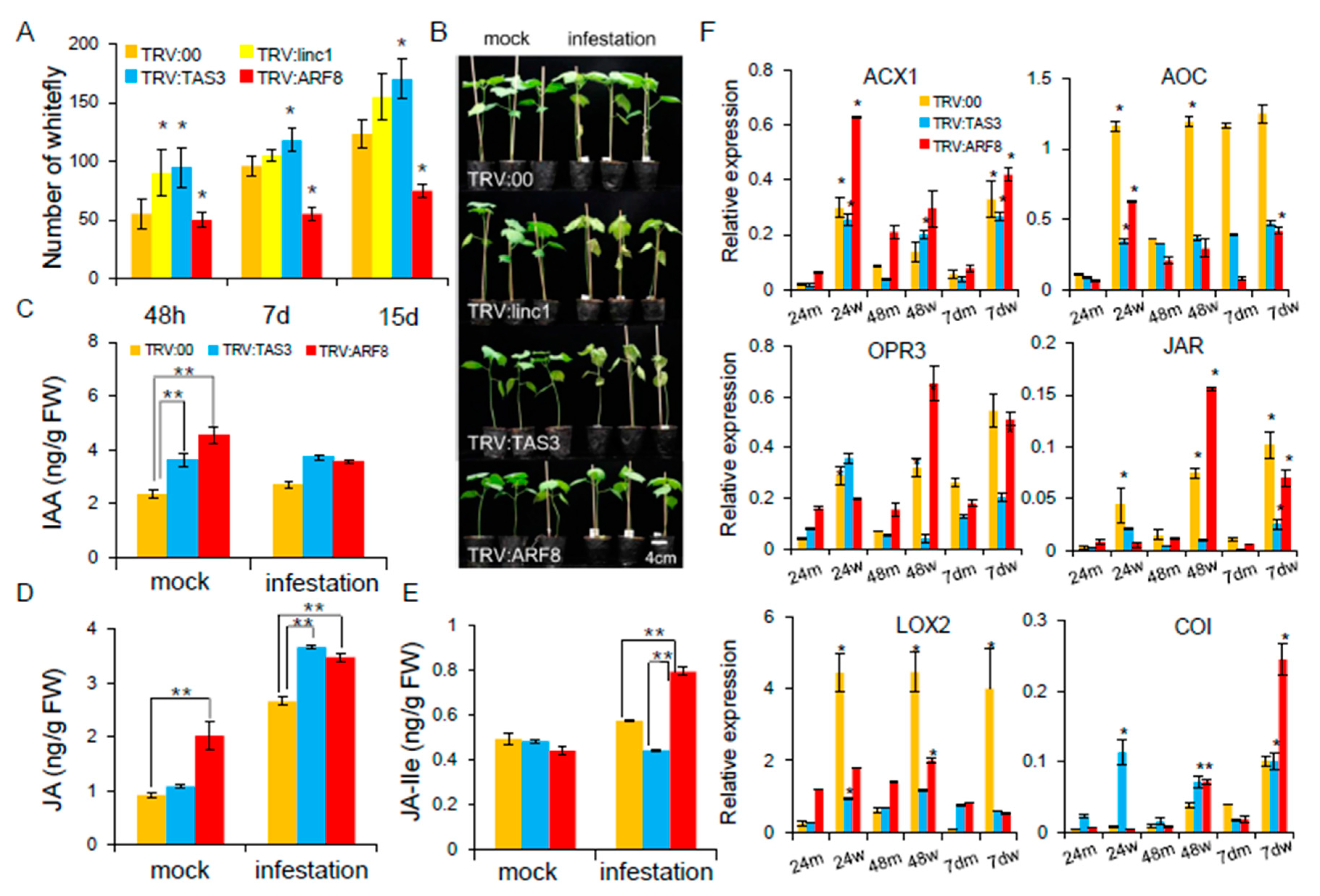
2.7. Characterization of the linc1-miR390-tasiARFs Cascade Involved in Cotton Response to Whitefly Infestation
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Whitefly Infestation, and RNA Isolation
4.2. Small RNA and Degradome Library Construction
4.3. miRNA Prediction Pipeline
4.4. Expression Profiles of miRNAs in the HR and ZS Plants during Whitefly Infestation
4.5. Identification and Functional Annotation of the miRNA Target Genes
4.6. Identification of lincRNAs from RNA-Seq Dataset
4.7. Stem-loop qRT-PCR Analysis of miRNAs
4.8. Identification of PHAS Loci
4.9. Validation of lincRNA Function and Corresponding Targets by VIGS
4.10. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, Z.J.; Scheffler, B.E.; Dennis, E.; Triplett, B.A.; Zhang, T.; Guo, W.; Chen, X.; Stelly, D.M.; Rabinowicz, P.D.; Town, C.D.; et al. Toward Sequencing Cotton (Gossypium) Genomes. Plant Physiol. 2007, 145, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wu, K.; Jiang, Y.; Xia, B.; Li, P.; Feng, H.; Wyckhuys, K.A.G.; Guo, Y. Mirid Bug Outbreaks in Multiple Crops Correlated with Wide-Scale Adoption of Bt Cotton in China. Science 2010, 328, 1151–1154. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Kanagaraj, A.; Verma, D.; Lange, T.; Daniell, H. Release of hormones from conjugates: Chloroplast expression of β-glucosidase results in elevated phytohormone levels associated with significant increase in biomass and protection from aphids or whiteflies conferred by sucrose esters. Plant Physiol. 2011, 155, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Zhang, X.; Daniell, H. Pinellia ternata agglutinin expression in chloroplasts confers broad spectrum resistance against aphid, whitefly, Lepidopteran insects, bacterial and viral pathogens. Plant Biotechnol. J. 2012, 10, 313–327. [Google Scholar] [CrossRef]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef]
- Shukla, A.K.; Upadhyay, S.K.; Mishra, M.; Saurabh, S.; Singh, R.; Singh, H.; Thakur, N.; Rai, P.; Pandey, P.; Hans, A.L.; et al. Expression of an insecticidal fern protein in cotton protects against whitefly. Nat. Biotechnol. 2016, 34, 1046–1051. [Google Scholar] [CrossRef]
- Zhu, L.; Li, J.; Xu, Z.; Manghwar, H.; Liang, S.; Li, S.; Alariqi, M.; Jin, S.; Zhang, X. Identification and selection of resistance to Bemisia tabaci among 550 cotton genotypes in the field and greenhouse experiments. Front. Agric. Sci. Eng. 2018, 5, 236–252. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Boil. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of Plant Small RNAs in Biotic Stress Responses. Annu. Rev. Plant Boil. 2009, 60, 485–510. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, M.; Gustafson, P.; Langridge, P.; Shi, B.J. Differential expression of microRNAs and other small RNAs in barley between water and drought conditions. Plant Biotechnol. J. 2015, 13, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-Directed Phasing during Trans-Acting siRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Ronemus, M.; Vaughn, M.W.; Martienssen, R.A. MicroRNA-Targeted and Small Interfering RNA–Mediated mRNA Degradation Is Regulated by Argonaute, Dicer, and RNA-Dependent RNA Polymerase in Arabidopsis. Plant Cell 2006, 18, 1559–1574. [Google Scholar] [CrossRef]
- Felippes, F.F.; Weigel, D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 2009, 10, 264–270. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Y.; Wu, J.; Ding, B.; Fei, Z. A dynamic evolutionary and functional landscape of plant phased small interfering RNAs. BMC Boil. 2015, 13, 32. [Google Scholar] [CrossRef]
- Zhai, J.; Jeong, D.-H.; De Paoli, E.; Park, S.; Rosen, B.D.; Li, Y.; González, A.J.; Yan, Z.; Kitto, S.L.; Grusak, M.A.; et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Wu, J.; Baldwin, I.T. New Insights into Plant Responses to the Attack from Insect Herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- Wu, J.; Hettenhausen, C.; Meldau, S.; Baldwin, I.T. Herbivory Rapidly Activates MAPK Signaling in Attacked and Unattacked Leaf Regions but Not between Leaves of Nicotiana attenuata. Plant Cell 2007, 19, 1096–1122. [Google Scholar] [CrossRef]
- Ellis, J.; Dodds, P.; Pryor, T. Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Boil. 2000, 3, 278–284. [Google Scholar] [CrossRef]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Boil. 2006, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Bozorov, T.A.; Baldwin, I.T.; Kim, S.-G. Identification and profiling of miRNAs during herbivory reveals jasmonate-dependent and -independent patterns of accumulation in Nicotiana attenuata. BMC Plant Boil. 2012, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Shahi, P.; Gase, K.; Baldwin, I.T. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc. Natl. Acad. Sci. USA 2008, 105, 4559–4564. [Google Scholar] [CrossRef]
- Sattar, S.; Addo-Quaye, C.; Thompson, G.A. miRNA-mediated auxin signalling repression during Vat-mediated aphid resistance in Cucumis melo. Plant Cell Environ. 2016, 39, 1216–1227. [Google Scholar] [CrossRef]
- Li, J.; Zhu, L.; Hull, J.J.; Liang, S.; Daniell, H.; Jin, S.; Zhang, X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insectBemisia tabaci(whitefly). Plant Biotechnol. J. 2016, 14, 1956–1975. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Fan, C.; Hao, Z.; Yan, J.; Li, G. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genom. 2015, 16, 793. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and Their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 Interaction and Dual Functionality in TAS3 Trans-Acting siRNA Formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef]
- Arikit, S.; Xia, R.; Kakrana, A.; Huang, K.; Zhai, J.; Yan, Z.; Valdés-López, O.; Prince, S.; Musket, T.A.; Nguyen, H.T.; et al. An Atlas of Soybean Small RNAs Identifies Phased siRNAs from Hundreds of Coding Genes. Plant Cell 2014, 26, 4584–4601. [Google Scholar] [CrossRef] [PubMed]
- Sattar, S.; Song, Y.; Anstead, J.A.; Sunkar, R.; Thompson, G.A. Cucumis meloMicroRNA Expression Profile During Aphid Herbivory in a Resistant and Susceptible Interaction. Mol. Plant-Microbe Interact. 2012, 25, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Kettles, G.J.; Drurey, C.; Schoonbeek, H.-J.; Maule, A.J.; Hogenhout, S.A. Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. New Phytol. 2013, 198, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Shao, Y.; Jiang, J.; Du, X.; Sheng, L.; Chen, F.; Fang, W.; Guan, Z.; Chen, S. MicroRNA Expression Profile during Aphid Feeding in Chrysanthemum (Chrysanthemum morifolium). PLoS ONE 2015, 10, e0143720. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, R.; Denholm, I.; Morin, S. Resistance to Insecticides in the TYLCV vector, Bemisia Tabaci. In Tomato Yellow Leaf Curl Virus Disease; Springer Science and Business Media LLC: Berlin, Germany, 2007; pp. 305–325. [Google Scholar]
- Wang, M.; Yuan, D.; Tu, L.; Gao, W.; He, Y.; Hu, H.; Wang, P.; Liu, N.; Lindsey, K.; Zhang, X. Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol. 2015, 207, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ju, Z.; Cao, D.; Zhai, B.; Qin, G.; Zhu, H.; Fu, D.; Luo, Y.; Zhu, B. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 2015, 13, 370–382. [Google Scholar] [CrossRef]
- Eckardt, N.A. A microRNA cascade in plant defense. Plant Cell 2012, 24, 840. [Google Scholar] [CrossRef]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef]
- Foyer, C.H.; Verrall, S.R.; Hancock, R.D. Systematic analysis of phloem-feeding insect-induced transcriptional reprogramming in Arabidopsis highlights common features and reveals distinct responses to specialist and generalist insects. J. Exp. Bot. 2015, 66, 495–512. [Google Scholar] [CrossRef]
- Plavskin, Y.; Nagashima, A.; Perroud, P.-F.; Hasebe, M.; Quatrano, R.S.; Atwal, G.S.; Timmermans, M.C. Ancient trans-Acting siRNAs Confer Robustness and Sensitivity onto the Auxin Response. Dev. Cell 2016, 36, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Gaquerel, E.; Stitz, M. Insect Resistance: An Emerging Molecular Framework Linking Plant Age and JA Signaling. Mol. Plant 2017, 10, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.; Zhang, X.; Liu, D.; Jin, S.; Cao, J.; Zhu, L.; Deng, F.; Tan, J.; Zhang, C. Suitable internal control genes for qRT-PCR normalization in cotton fiber development and somatic embryogenesis. Chin. Sci. Bull. 2007, 52, 3110–3117. [Google Scholar] [CrossRef]
- German, M.A.; Pillay, M.; Jeong, D.-H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational Identification of Plant MicroRNAs and Their Targets, Including a Stress-Induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Yang, X.; Li, L. miRDeep-P: A computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011, 27, 2614–2615. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Boil. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef]
- Guo, Q.; Qu, X.; Jin, W. PhaseTank: Genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics 2015, 31, 284–286. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Xiao, J.; Wang, S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: Application in study of rice-bacterium interaction. Plant Methods 2012, 8, 2. [Google Scholar] [CrossRef]
| Sample | Raw Reads | Unique Reads | SnoRNA | snRNA | 5S_rRNA | sRNA | Mapping |
|---|---|---|---|---|---|---|---|
| HR0_R1 | 16,906,187 | 2,276,889 | 567 | 333 | 6957 | 977,245 | 84.19% |
| HR0_R2 | 21,306,455 | 2,281,508 | 949 | 554 | 10,990 | 600,015 | 83.81% |
| HR0_R3 | 11,613,784 | 729,752 | 213 | 227 | 2721 | 650,351 | 84.12% |
| HR24_R1 | 20,313,659 | 2,350,845 | 548 | 394 | 7678 | 824,200 | 81.82% |
| HR24_R2 | 22,215,560 | 2,404,771 | 184 | 93 | 1669 | 1,476,308 | 82.16% |
| HR24_R3 | 11,306,605 | 833,843 | 245 | 262 | 2932 | 745,572 | 84.45% |
| ZS0_R1 | 18,328,323 | 1,838,613 | 97 | 43 | 770 | 1,047,368 | 79.65% |
| ZS0_R2 | 21,173,195 | 2,457,802 | 149 | 68 | 1480 | 1,476,875 | 79.93% |
| ZS0_R3 | 11,335,617 | 1,086,277 | 232 | 245 | 2977 | 986,067 | 84.21% |
| ZS24_R1 | 18,240,711 | 1,104,283 | 70 | 44 | 940 | 636,559 | 81.41% |
| ZS24_R2 | 17,720,064 | 1,837,592 | 108 | 69 | 1051 | 507,170 | 82.14% |
| ZS24_R3 | 11,416,722 | 1,037,485 | 220 | 232 | 3176 | 925,213 | 83.51% |
| MiRNA_ID | miRBase21 | Strand | Chr | Start | End | lincRNA_ID | miRNA Target Annotation |
|---|---|---|---|---|---|---|---|
| P132 | osa-miR171f-3p | + | A05 | 15999334 | 15999553 | GhA05linc.520 | GRAS |
| P147 | ghr-miR166b | + | A07 | 28328142 | 28328522 | GhA07linc.319 | Homeobox-leucine zipper |
| P147 | ghr-miR166b | + | A08 | 8999568 | 8999969 | GhA08linc.292 | Homeobox-leucine zipper |
| P147 | ghr-miR166b | + | A11 | 67055538 | 67055606 | GhA11linc.93 | Homeobox-leucine zipper |
| P149 | NoHits | + | A08 | 103041407 | 103042061 | GhA08linc.135 | |
| P168 | gra-miR8733 | - | D06 | 692582 | 692872 | GhD06linc.129 | |
| P181 | ath-miR172b-3p | - | A05 | 9079609 | 9079903 | GhA05linc.451 (linc6) | related to AP2.7 |
| P187 | ghr-miR390c | + | D09 | 43446164 | 43448093 | GhD09linc.75 (linc1) | TAS3 |
| P193 | ghr-miR156d | + | A07 | 2556261 | 2556647 | GhA07linc.14 | SPL |
| P72 | gra-miR482c | + | A07 | 9733996 | 9734022 | GhA07linc.38 (linc4) | NB-ARC |
| P73 | NoHits | + | D05 | 43103308 | 43103635 | GhD05linc.279 (linc5) | NB-ARC |
| P73 | NoHits | - | D05 | 43102918 | 43103615 | GhD05linc.670 (linc2) | NB-ARC |
| P81 | NoHits | + | D05 | 43103308 | 43103635 | GhD05linc.279 | NB-ARC |
| P81 | NoHits | - | D05 | 43102918 | 43103615 | GhD05linc.670 | NB-ARC |
| P87 | NoHits | + | A12 | 84153544 | 84153795 | GhA12linc.146 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Hull, J.J.; Liang, S.; Wang, Q.; Chen, L.; Zhang, Q.; Wang, M.; Mansoor, S.; Zhang, X.; Jin, S. Genome-Wide Analysis of Cotton miRNAs During Whitefly Infestation Offers New Insights into Plant-Herbivore Interaction. Int. J. Mol. Sci. 2019, 20, 5357. https://doi.org/10.3390/ijms20215357
Li J, Hull JJ, Liang S, Wang Q, Chen L, Zhang Q, Wang M, Mansoor S, Zhang X, Jin S. Genome-Wide Analysis of Cotton miRNAs During Whitefly Infestation Offers New Insights into Plant-Herbivore Interaction. International Journal of Molecular Sciences. 2019; 20(21):5357. https://doi.org/10.3390/ijms20215357
Chicago/Turabian StyleLi, Jianying, J. Joe Hull, Sijia Liang, Qiongqiong Wang, Luo Chen, Qinghua Zhang, Maojun Wang, Shahid Mansoor, Xianlong Zhang, and Shuangxia Jin. 2019. "Genome-Wide Analysis of Cotton miRNAs During Whitefly Infestation Offers New Insights into Plant-Herbivore Interaction" International Journal of Molecular Sciences 20, no. 21: 5357. https://doi.org/10.3390/ijms20215357
APA StyleLi, J., Hull, J. J., Liang, S., Wang, Q., Chen, L., Zhang, Q., Wang, M., Mansoor, S., Zhang, X., & Jin, S. (2019). Genome-Wide Analysis of Cotton miRNAs During Whitefly Infestation Offers New Insights into Plant-Herbivore Interaction. International Journal of Molecular Sciences, 20(21), 5357. https://doi.org/10.3390/ijms20215357







