Overexpression of OsPIN2 Regulates Root Growth and Formation in Response to Phosphate Deficiency in Rice
Abstract
:1. Introduction
2. Results
2.1. The Expression of OsPIN2 Was Induced by Low phosphate (LP) Supply
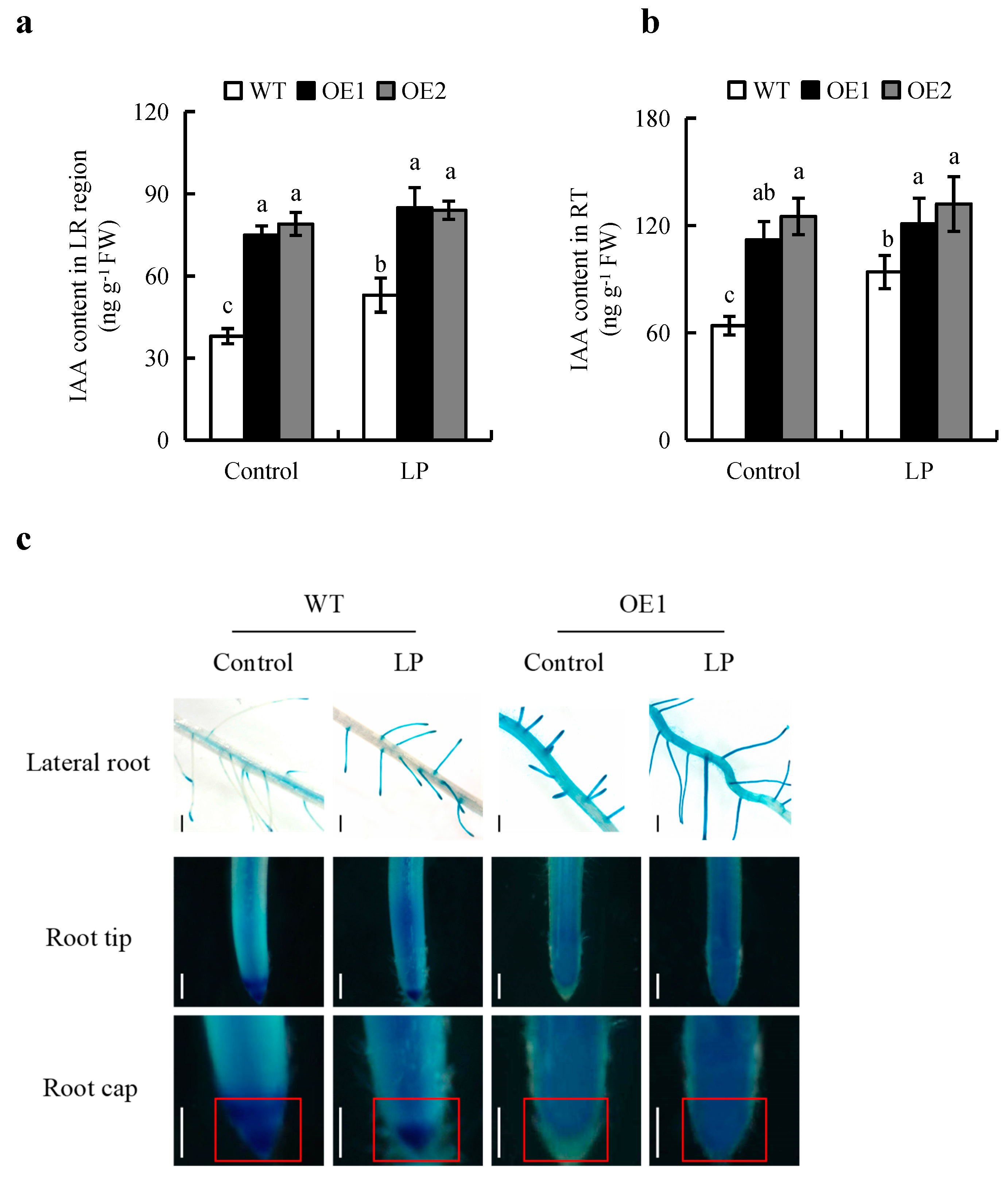
2.2. Overexpression of OsPIN2 Regulates Auxin Levels in Roots
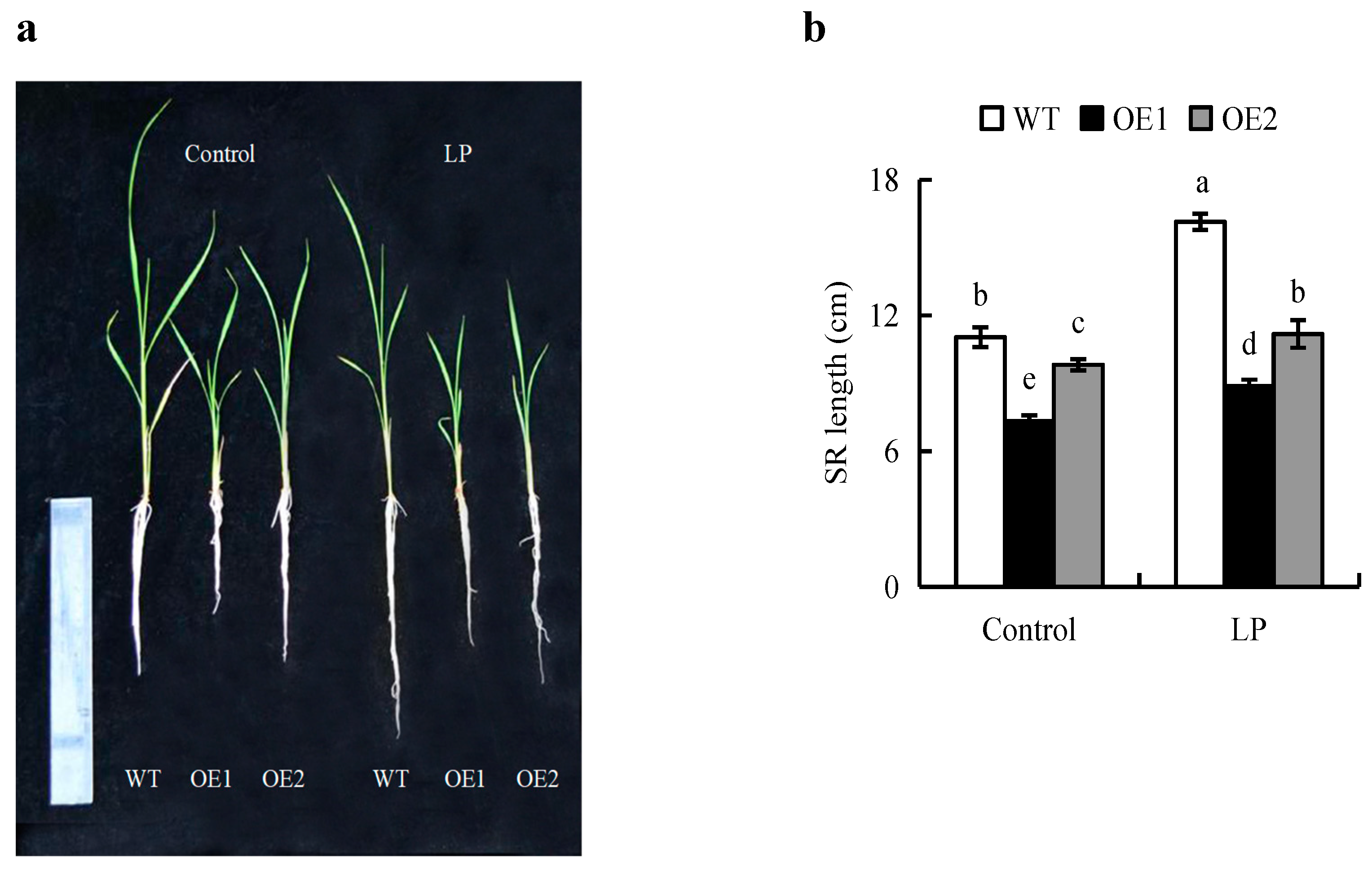
2.3. The Root Architectures of OE Lines Were Less Sensitive Than WT Plants to LP Condition
2.4. Overexpressing OsPIN2 Regulates Root Development by Modifying Auxin Distribute
3. Discussion
3.1. Overexpression of OsPIN2 Affects Root Elongation by Regulating the Elongation of Epidermal Cells that Leave the Root Meristem
3.2. Overexpression of OsPIN2 Regulate RHs and LRs Formation via Change Auxin Distribute in Root Transversal Section
4. Materials and Methods
4.1. Plant Materials
4.2. Plant Growth
4.3. Measurement of Root System Architecture
4.4. pPIN2::GUS Construct
4.5. Determination of IAA Content
4.6. [3H]IAA Transport
4.7. Cortical Cell Length Analysis
4.8. pCYCB1;1::GUS Construct
4.9. Quantitative Reverse-Transcription Polymerase Chain Reaction Analyses
4.10. Data Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forde, B.; Lorenzo, H. The nutritional control of root development. Plant Soil. 2001, 232, 51–68. [Google Scholar] [CrossRef]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, S.; Wang, L.; Yang, Y.; Zhang, H.; Cui, H.; Shao, H.; Xu, G. OsPht1;8, a phosphate transporter, is involved in auxin and phosphate starvation response in rice. J. Exp. Bot. 2017, 68, 5057–5068. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tao, J.; Bi, Y.; Xie, X.; Yoneyama, K.; Zhao, Q.; Xu, G.; Zhang, Y. OsPIN1b is involved in rice seminal root elongation by regulating root apical meristem activity in response to nitrogen- and phosphate-deficiency. Sci. Rep. 2018, 8, 13014. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef]
- Kellermeier, F.; Armengaud, P.; Seditas, T.J.; Danku, J.; Salt, D.E.; Amtmann, A. Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 2014, 26, 1480–1496. [Google Scholar] [CrossRef]
- Péret, B.; Desnos, T.; Jost, R.; Kanno, S.; Berkowitz, O.; Nussaume, L. Root architecture responses: In search of phosphate. Plant Physiol. 2014, 166, 1713–1723. [Google Scholar] [CrossRef]
- Sun, H.; Bi, Y.; Tao, J.; Huang, S.; Hou, M.; Xue, R.; Liang, Z.; Xie, X.; Yoneyama, K.; Shen, Q.; et al. Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiency in rice. Plant Cell Environ. 2016, 39, 1473–1484. [Google Scholar] [CrossRef]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef]
- Sun, H.; Tao, J.; Liu, S.; Huang, S.; Chen, S.; Xie, X.; Yoneyama, K.; Zhang, Y.; Xu, G. Strigolactones are involved in phosphate and nitrate deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef]
- Giri, J.; Bhosale, R.; Huang, G.; Pandey, B.K.; Parker, H.; Zappala, S.; Yang, J.; Dievart, A.; Bureau, C.; Ljung, K.; et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018, 9, 1408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Simpson, R.; Kim, C.; Warthmann, N.; Delhaize, E.; Dolan, L.; Byrne, M.; Wu, Y.; Ryan, P. Do longer root hairs improve phosphorus uptake? Testing the hypothesis with transgenic Brachypodium distachyon lines overexpressing endogenous RSL genes. New Phytol. 2018, 217, 1654–1666. [Google Scholar] [CrossRef] [PubMed]
- Inahashia, H.; Shelleyb, I.; Yamauchia, T.; Nishiuchia, S.; Takahashi-Nosakab, M.; Matsunamie, M.; Ogawa, A.; Noda, Y.; Inukai, Y. OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant. 2018, 164, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, M.; Li, Y.; Ruan, W.; Mo, X.; Wu, Z.; Sturrock, C.; Yu, H.; Lu, C.; Peng, J.; et al. LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot. 2018, 69, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Wang, S.; Zhang, S.; Xu, Y.; Qian, Q.; Qi, Y.; Jiang, D. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Bhalerao, R.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011, 23, 3961–3973. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant. 2012, 5, 334–338. [Google Scholar] [CrossRef]
- Friml, J. Auxin transport-shaping the plant. Curr. Opin. Plant Biol. 2003, 6, 712. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef]
- Zazimalova, E.; Murphy, A.; Yang, H.; Hoyerova, K.; Hosek, P. Auxin transporters-why so many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Guan, C.; Gälweiler, L.; Tänzler, P.; Huijser, P.; Marchant, A.; Parry, G.; Bennett, M.; Wisman, E.; Palme, K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998, 17, 6903–6911. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Shin, H.S.; Guo, Z.; Blancaflor, E.B.; Masson, P.H.; Chen, R. Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J. 2005, 42, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, J.; Xu, J.; Seifertová, D.; Brewer, P.; Růžička, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Feng, Q.; Zhang, Y.; Hao, P.; Wang, S.; Fu, G.; Huang, Y.; Li, Y.; Zhu, J.; Liu, Y.; Hu, X.; et al. Sequence and analysis of rice chromosome 4. Nature 2002, 420, 316–320. [Google Scholar] [CrossRef]
- Sasaki, T.; Matsumoto, T.; Yamamoto, K. The genome sequence and structure of rice chromosome 1. Nature 2002, 420, 312–316. [Google Scholar] [CrossRef]
- Wang, J.; Hu, H.; Wang, G.; Li, J.; Chen, J.; Wu, P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant. 2009, 2, 823–831. [Google Scholar] [CrossRef]
- Scheres, B.; Benfey, P.; Dolan, L. Root devepopment. Arab. Book 2002, 1, e0101. [Google Scholar] [CrossRef]
- Blakeslee, J.; Peer, W.; Murphy, A. Auxin transport. Curr. Opin. Plant Biol. 2005, 8, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Cho, H. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 2013, 4, 448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Li, C.; Wu, Z.; Jia, Y.; Wang, H.; Sun, S.; Mao, C.; Wang, X. Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 2017, 8, 1121. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Cho, L.-H.; Kim, Y.-J.; Gho, Y.-S.; Jeong, H.Y.; Hong, W.-J.; Lee, C.; Park, H.; Jwa, N.-S.; Dangol, S.; et al. RSL class II transcription factors guide the nuclear localization of RHL1 to regulate root hair development. Plant Physiol. 2018, 179, 558–568. [Google Scholar] [CrossRef] [PubMed]
- Muday, G.K.; Rahman, A. Auxin transport and the integration of gravitropic growth. Plant Trop. 2008, 3, 47–78. [Google Scholar]
- Feraru, E.; Friml, J. PIN polar targeting. Plant Physiol. 2008, 147, 1553–1559. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, J.; Song, W.; Tao, J.; Huang, S.; Chen, S.; Hou, M.; Xu, G.; Zhang, Y. Nitric oxide generated by nitrate reductase increases nitrogen uptake capacity by inducing lateral root formation and inorganic nitrogen uptake under partial nitrate nutrition in rice. J. Exp. Bot. 2015, 66, 2449–2459. [Google Scholar] [CrossRef] [Green Version]
- Malamy, J.; Benfey, P. Organization and cell differentiation in lateral root Arabidopsis thaliana. Development 1997, 124, 33–44. [Google Scholar]
- Pemberton, L.; Tsai, S.; Lovell, P.; Harris, P. Epidermal patterning in seedling roots of eudicotyledons. Ann. Bot. 2001, 87, 649–654. [Google Scholar] [CrossRef]
- Wu, D.; Shen, H.; Yokawa, K.; Baluška, F. Overexpressing OsPIN2 enhances aluminium internalization by elevating vesicular trafficking in rice root apex. J. Exp. Bot. 2015, 66, 6791–6801. [Google Scholar] [CrossRef]
- Sun, S.; Gu, M.; Cao, Y.; Huang, X.; Zhang, X.; Ai, P.; Zhao, J.; Fan, X.; Xu, G. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.; Li, J.; Gong, X.; Huang, S.; Zhu, X.; Zhang, Y.; Xu, G. Auxin distribution is differentially affected by nitrate in roots of two rice cultivars differing in responsiveness to nitrogen nutrients. Ann. Bot. 2013, 112, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; An, X.; Cheng, L.; Chen, F.; Bao, J.; Yuan, L.; Zhang, F.; Mi, G. Auxin transport in maize roots in response to localized nitrate supply. Ann. Bot. 2010, 106, 1019–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, L.; Zhang, B.; Mao, C.; Li, J.; Wu, Y.; Wu, P.; Wu, Z. OsCYTINV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.). Planta 2008, 228, 51–59. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Guo, X.; Xu, F.; Wu, D.; Zhang, X.; Lou, M.; Luo, F.; Xu, G.; Zhang, Y. Overexpression of OsPIN2 Regulates Root Growth and Formation in Response to Phosphate Deficiency in Rice. Int. J. Mol. Sci. 2019, 20, 5144. https://doi.org/10.3390/ijms20205144
Sun H, Guo X, Xu F, Wu D, Zhang X, Lou M, Luo F, Xu G, Zhang Y. Overexpression of OsPIN2 Regulates Root Growth and Formation in Response to Phosphate Deficiency in Rice. International Journal of Molecular Sciences. 2019; 20(20):5144. https://doi.org/10.3390/ijms20205144
Chicago/Turabian StyleSun, Huwei, Xiaoli Guo, Fugui Xu, Daxia Wu, Xuhong Zhang, Manman Lou, Feifei Luo, Guohua Xu, and Yali Zhang. 2019. "Overexpression of OsPIN2 Regulates Root Growth and Formation in Response to Phosphate Deficiency in Rice" International Journal of Molecular Sciences 20, no. 20: 5144. https://doi.org/10.3390/ijms20205144



