Genome-Wide Analysis of Dynamin Gene Family in cassava (Manihot esculenta Crantz) and Transcriptional Regulation of Family Members ARC5 in Hormonal Treatments
Abstract
:1. Introduction
2. Results
2.1. Identification of the Dynamin Proteins in Cassava
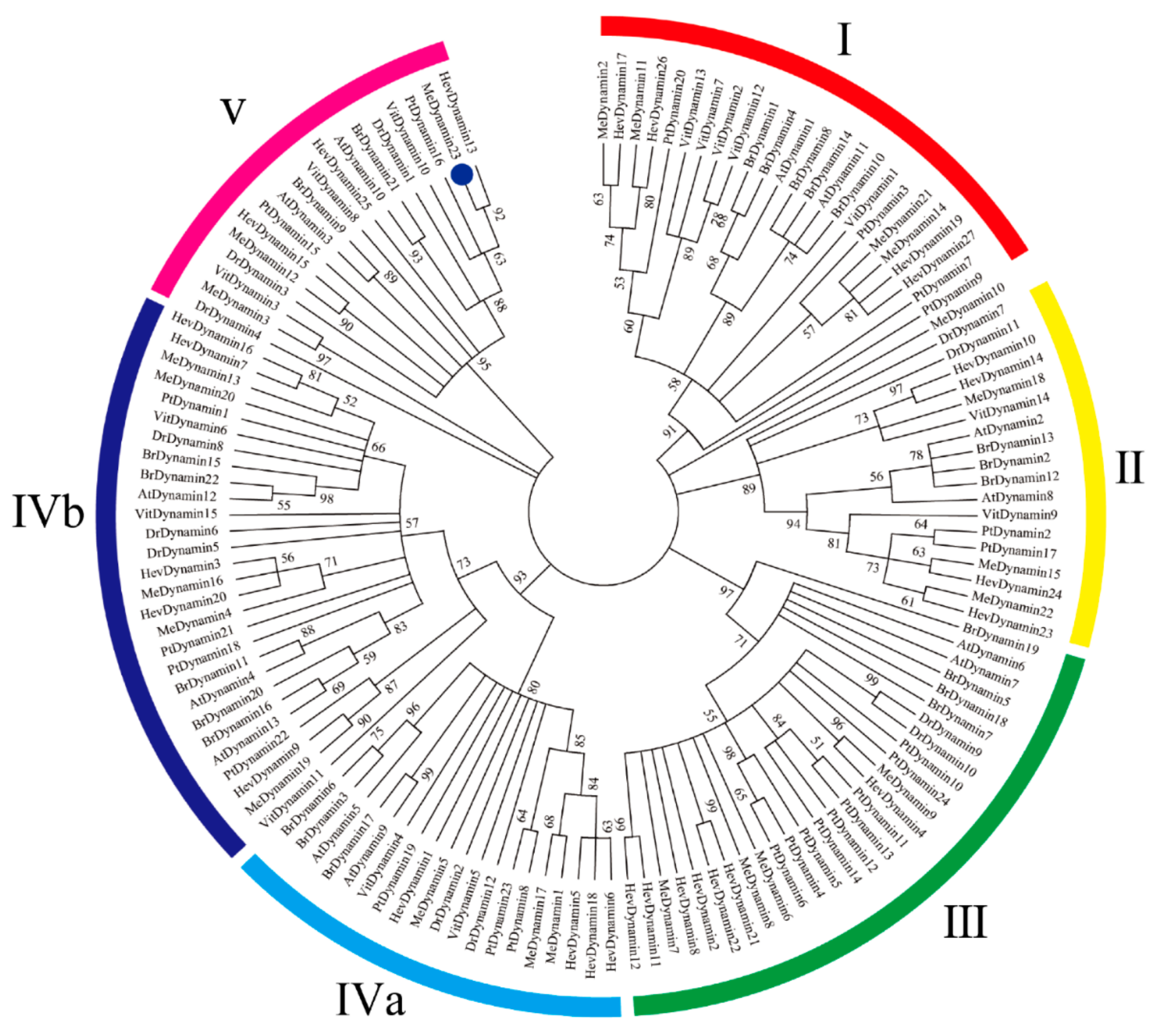
2.2. Multiple Sequence Alignment, Phylogenetic Analysis, and Classification of MeDynamins
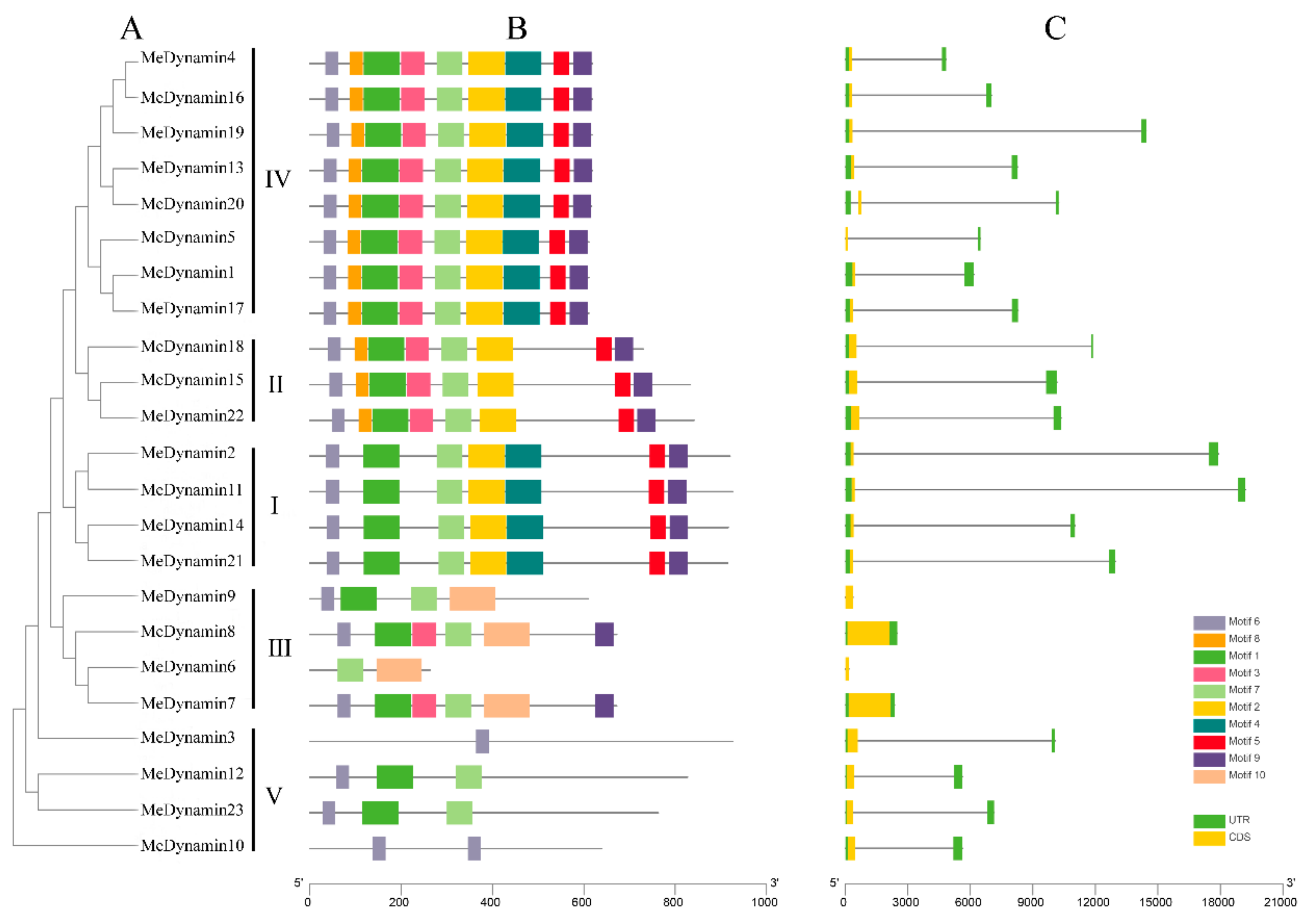
2.3. Gene Structure Features of MeDynamins
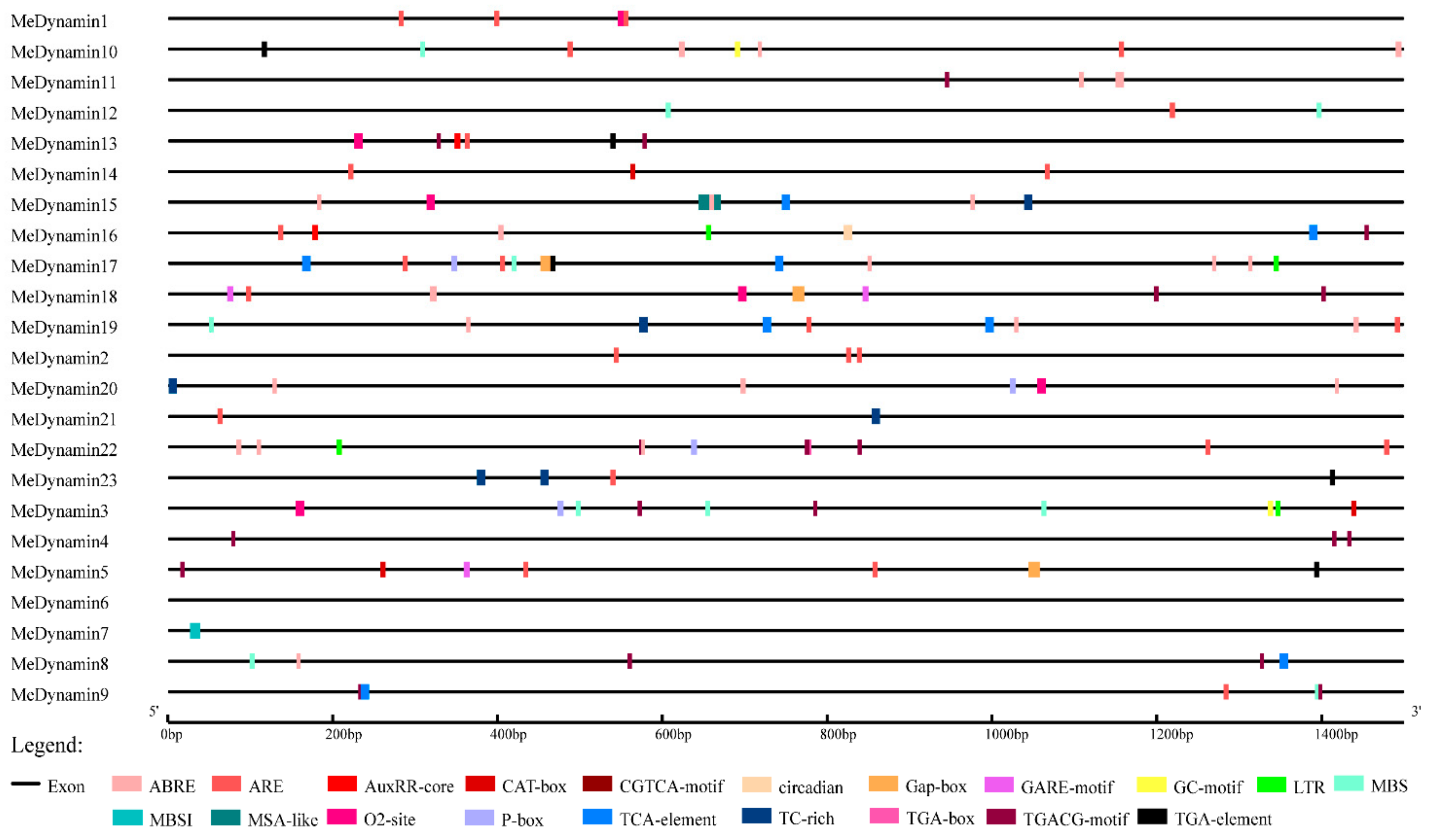
2.4. Promoter Analysis
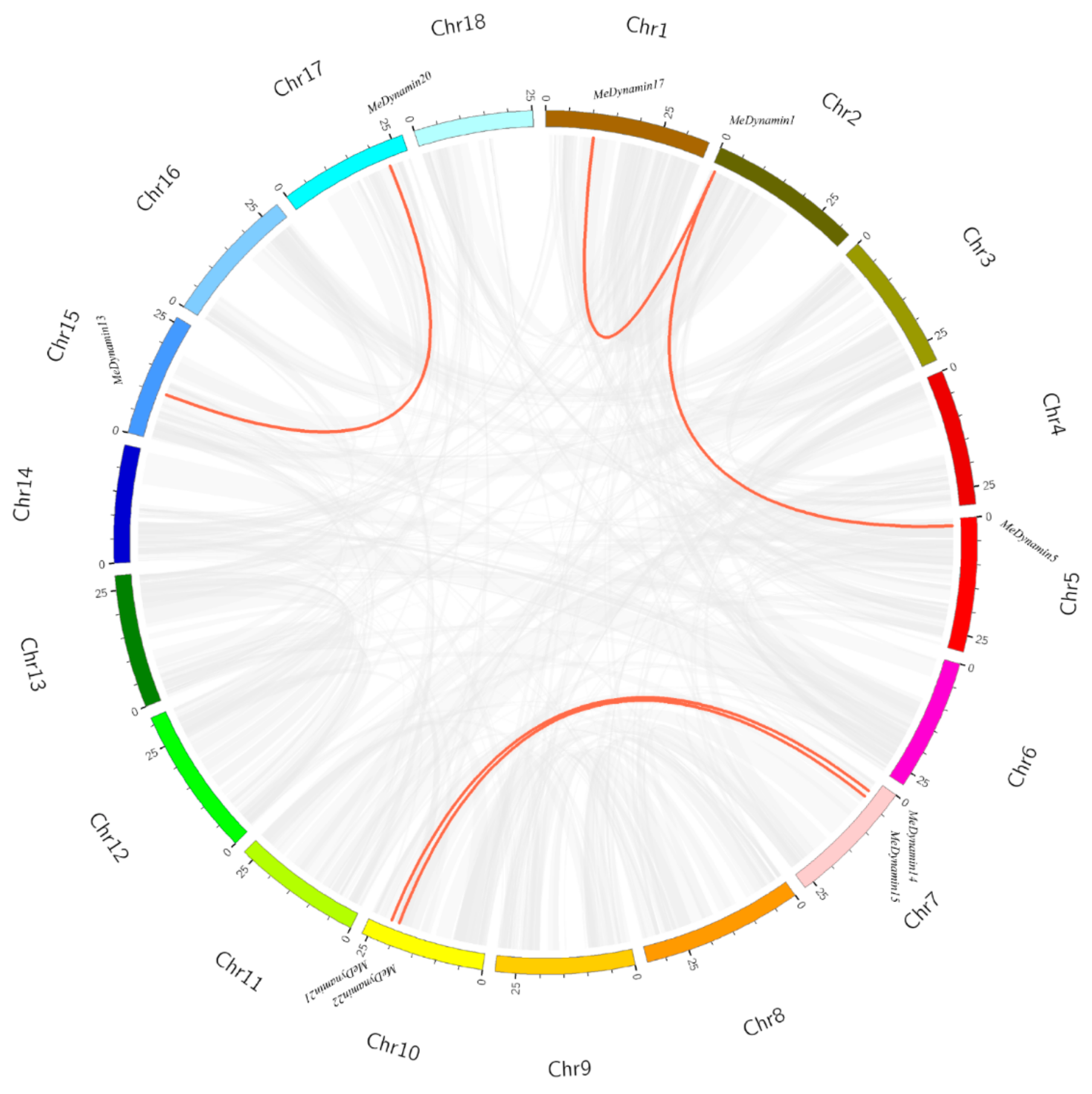
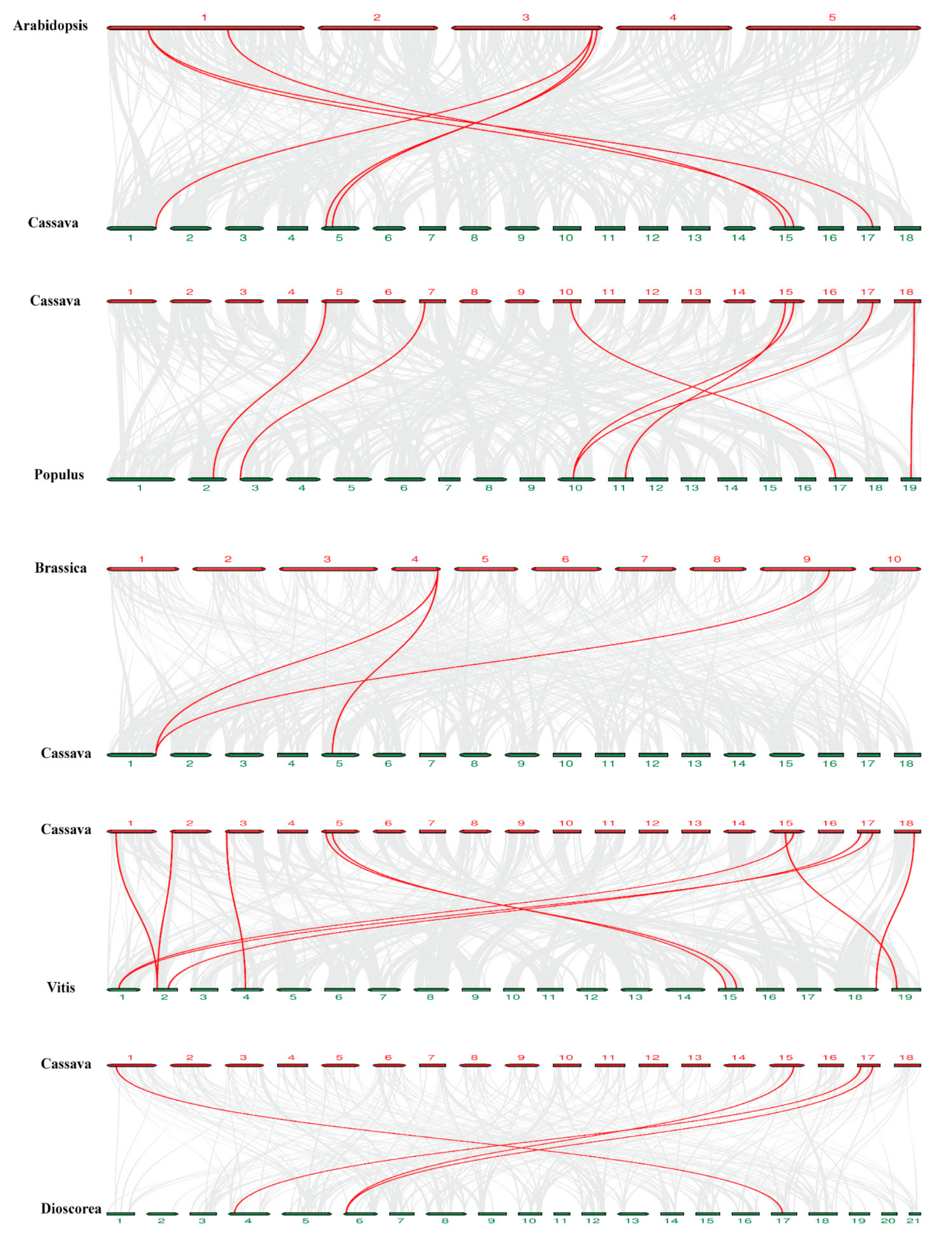
2.5. Chromosome Distribution and Synteny Analysis of MeDynamins Gene
2.6. Expression Patterns of MeDynamins in Different Tissues and Developmental Stages
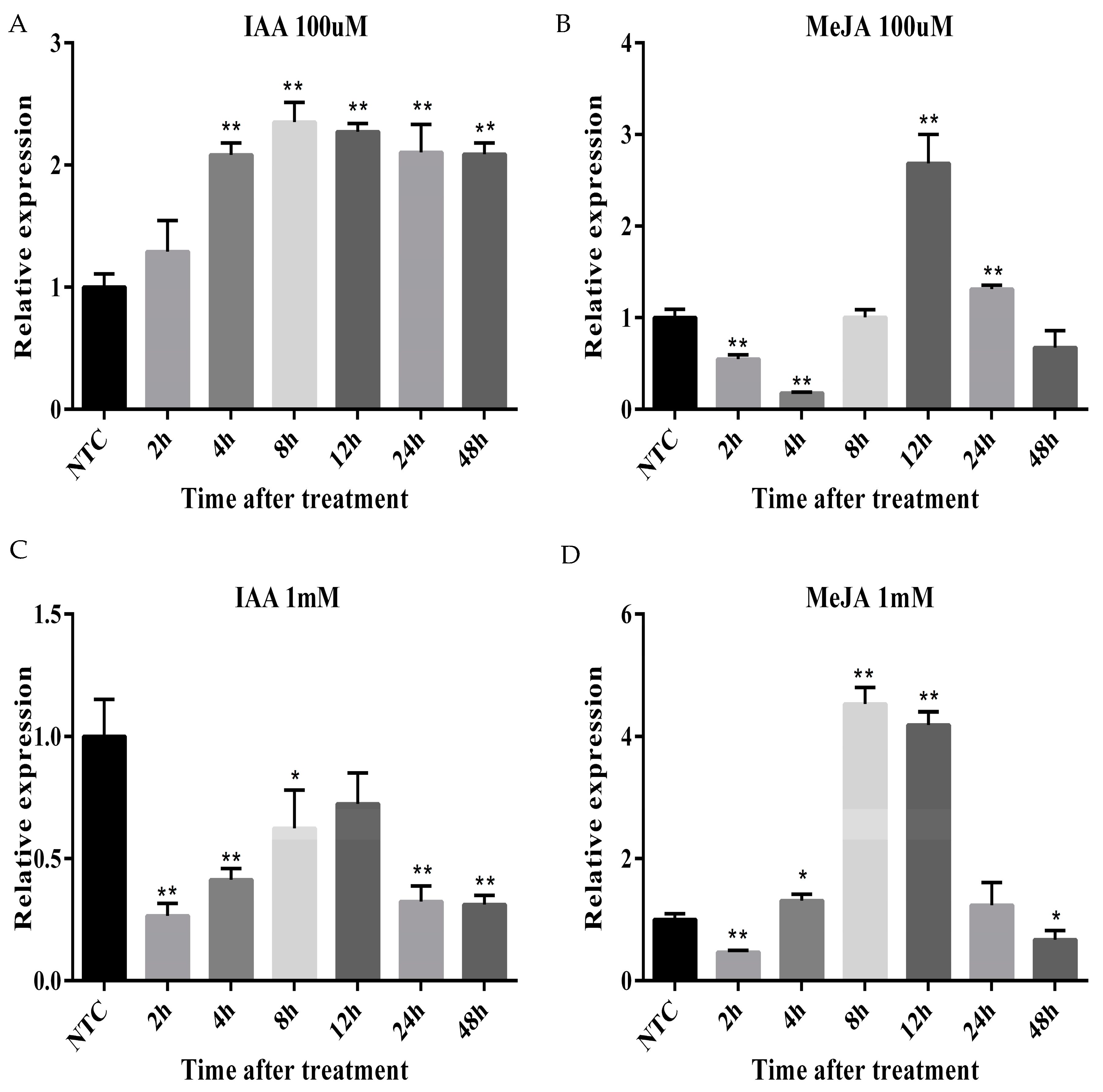
2.7. Differential Expression of ARC5 Gene under IAA and MeJA Treatments
3. Discussion
4. Materials and Methods
4.1. Identification of Dynamin Genes
4.2. Sequence Analysis and Structural Characterization
4.3. Chromosomal localization and Gene Duplication
4.4. Phylogenetic Analysis of Cassava Dynamin Genes Family
4.5. Analysis of Cis-Elements in Promoter Region
4.6. RNA-Sequencing (RNA-seq) Data Analysis of Dynamin Genes
4.7. Plant Materials
4.8. RNA Extraction and qRT-PCR Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Cavalier-Smith, T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000, 5, 174–182. [Google Scholar] [CrossRef]
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Stokes, K.D.; Rutherford, S.M.; Percival, A.L.; Lee, W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 1998, 10, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S. ARC6 Is a J-Domain Plastid Division Protein and an Evolutionary Descendant of the Cyanobacterial Cell Division Protein Ftn2. Plant Cell Online 2003, 15, 1918–1933. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Froehlich, J.E.; Osteryoung, K.W. PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 2006, 18, 2517–2530. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Kadirjan-Kalbach, D.; Froehlich, J.E.; Osteryoung, K.W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 2003, 100, 4328–4333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyagishima, S.Y.; Wolk, C.P.; Osteryoung, K.W. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 2005, 56, 126–143. [Google Scholar] [CrossRef]
- Shimida, H.; Koizumi, M.; Kuroki, K.; Mochizuki, M.; Fujimoto, H.; Ohta, H.; Masuda, T.; Takamiya, K. ARC3, a Chloroplast Division Factor, is a Chimera of Prokaryotic FtsZ and Part of Eukaryotic Phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 2004, 45, 960–967. [Google Scholar] [CrossRef]
- Raynaud, C.; Perennes, C.; Reuzeau, C.; Catrice, O.; Brown, S.; Bergounioux, C. Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc. Natl. Acad. Sci. USA 2005, 102, 8216–8221. [Google Scholar] [CrossRef] [Green Version]
- Haswell, E.S.; Meyerowitz, E.M. MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 2006, 16, 1–11. [Google Scholar] [CrossRef]
- Hong, Z.; Geisler-Lee, C.J.; Zhang, Z.; Verma, D.P.S. Phragmoplastin dynamics: Multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol. Biol. 2003, 53, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.T.; Yasuzawa, M.; Kojo, K.H.; Niwa, Y.; Abe, T.; Yoshida, S.; Nakano, T.; Itoh, R.D. The Arabidopsis arc5 and arc6 mutations differentially affect plastid morphology in pavement and guard cells in the leaf epidermis. PLoS ONE 2018, 13, e0192380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, J. The Arabidopsis chloroplast division protein DYNAMIN-RELATED PROTEIN5B also mediates peroxisome division. Plant Cell 2011, 22, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, E.; Takechi, K.; Sato, H.; Yamada, T.; Takio, S.; Takano, H. Three dynamin-related protein 5B genes are related to plastid division in Physcomitrella patens. Plant Sci. 2011, 180, 789–795. [Google Scholar] [CrossRef]
- Shen, X.Z.; Liu, H.; Liu, X.Q.; Yuan, G.X.; Gao, Y.F.; Gao, H.B. Identification and Analysis of Mutant Gene ARC5 in Chloroplast Mutation Mutant cpd4. J. Plant Physiol. 2013, 49, 477–484. [Google Scholar]
- Pan, R.; Hu, J. The conserved fission complex on peroxisomes and mitochondria. Plant Signal. Behav. 2011, 6, 870–872. [Google Scholar] [CrossRef] [Green Version]
- Kang, B.H. Members of the Arabidopsis Dynamin-Like Gene Family, ADL1, Are Essential for Plant Cytokinesis and Polarized Cell Growth. Plant Cell Online 2003, 15, 899–913. [Google Scholar] [CrossRef]
- Glynn, J.M.; Miyagishima, S.Y.; Yoder, D.W.; Osteryoung, K.W.; Vitha, S. Chloroplast Division. Traffic 2007, 8, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Ungewickell, E.J.; Hinrichsen, L. Endocytosis: Clathrin-mediated membrane budding. Curr. Opin. Cell Biol. 2007, 19, 417–425. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, J. Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 2010, 57, 146–159. [Google Scholar] [CrossRef]
- Hoppins, S.; Lackner, L.; Nunnari, J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007, 76, 751–780. [Google Scholar] [CrossRef]
- Sever, S. Dynamin and endocytosis. Curr. Opin. Cell Biol. 2002, 14, 463–467. [Google Scholar] [CrossRef]
- Kar, U.P.; Dey, H.; Rahaman, A. Regulation of dynamin family proteins by post-translational modifications. J. Biosci. 2017, 42, 333–344. [Google Scholar] [CrossRef]
- Elsharkawy, M.A. Cassava biology and physiology. Plant Mol. Biol. 2004, 56, 481–501. [Google Scholar] [CrossRef] [PubMed]
- Nassar, N. Cassava, Manihot esculenta Crantz and wild relatives: Their relationships and evolution. Genet. Resour. Crop Evol. 2001, 48, 429–436. [Google Scholar] [CrossRef]
- Nweke, F.I.; Spencer, D.S.C.; Lynam, J.K. The Cassava Transformation: Africa’s Best-Kept Secret; Michigan State University Press: East Lansing, MI, USA, 2002. [Google Scholar]
- Jansson, C.; Westerbergh, A.; Zhang, J.; Hu, X.; Sun, C. Cassava, a potential biofuel crop in (the) People’s Republic of China. Appl. Energy 2009, 86 (Suppl. 1), S95–S99. [Google Scholar] [CrossRef]
- Nuwamanya, E.; Chiwona-Karltun, L.; Kawuki, R.S.; Baguma, Y. Bio-ethanol production from non-food parts of cassava (Manihot esculenta Crantz). Ambio 2012, 41, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.R.D.; Lacerda, L.G.; Bannach, G.; Schnitzler, E. Thermoanalytical study of native cassava starch treated with hydrogen peroxide. Aliment. E Nutr. 2011, 22, 7–15. [Google Scholar]
- Beninca, C.; Demiate, I.M.; Lacerda, L.G.; Carvalho Filho, M.A.S.; Ionashiro, M.; Schnitzler, E. Thermal behavior of corn starch granules modified by acid treatment at 30 and 50 °C. Eclética Química 2008, 33, 13–18. [Google Scholar] [CrossRef]
- Lacerda, L.G.; Filho, M.A.D.S.C.; Demiate, I.M.; Bannach, G.; Ionashiro, M.; Schnitzler, E. Thermal behaviour of corn starch granules under action of fungal α-amylase. J. Therm. Anal. Calorim. 2008, 93, 445–449. [Google Scholar] [CrossRef]
- Cui, Y.Y.; Tian, Z.M.; LI, Z.M.; Ma, X.Y. Nutritional Value of Cassava and Its By-products and Their Application in Animal Production. China Anim. Husb. Vet. Med. 2018, 49, 2135–2146. [Google Scholar]
- Wilson, M.C.; Mutka, A.M.; Hummel, A.W.; Berry, J.; Chauhan, R.D.; Vijayaraghavan, A.; Taylor, N.J.; Voytas, D.F.; Chitwood, D.H.; Bart, R.S. Gene expression atlas for the food security crop cassava. New Phytol. 2017, 4, 213. [Google Scholar] [CrossRef] [PubMed]
- Jilly, R.; Khan, N.Z.; Aronsson, H.; Schneider, D. Dynamin-Like Proteins Are Potentially Involved in Membrane Dynamics within Chloroplasts and Cyanobacteria. Front. Plant Sci. 2018, 9, 206. [Google Scholar] [CrossRef]
- Mun, J.H.; Yu, H.J.; Park, S.; Park, B.S. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol. Genet. Genom. 2009, 282, 617–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Wang, Y.; Chai, F.; Li, S.; Xin, H.; Liang, Z. Genome-wide identification and characterization of the 14-3-3 family in Vitis vinifera L. during berry development and cold- and heat-stress response. BMC Genom. 2018, 19, 579. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Guo, D.; Li, H.-L.; Zhu, J.-H.; Wang, Y.; An, F.; Xie, G.-S.; Peng, S.-Q. Genome-wide identification, characterization, and expression analysis of SnRK2 family in Hevea brasiliensis. Tree Genet Genomes 2017, 13, 86. [Google Scholar] [CrossRef]
- Wu, L.; Wang, B.; Yang, J.; Song, C.; Wang, P.; Chen, S.; Sun, W. The chloroplast genome sequence of an important medicinal plant Dioscorea nipponica. Mitochondrial DNA 2016, 27, 2559–2560. [Google Scholar]
- Brand, L.H.; Fischer, N.M.; Harter, K.; Kohlbacher, O.; Wanke, D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013, 41, 9764–9778. [Google Scholar] [CrossRef]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef]
- Ross, C.A.; Shen, Q.J.; Liu, Y. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2010, 49, 827–842. [Google Scholar] [CrossRef]
- Chothia, C.; Gough, J.; Vogel, C.; Teichmann, S.A. Evolution of the protein repertoire. Science 2003, 300, 1701–1703. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals by gene duplication. Hereditas 1968, 59, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Miyagishima, S.Y. Mechanism of plastid division: From a bacterium to an organelle. Plant Physiol. 2011, 155, 1533–1544. [Google Scholar] [CrossRef]
- Huang, J.; Fujimoto, M.; Fujiwara, M.; Fukao, Y.; Arimura, S.; Tsutsumi, N. Arabidopsis dynamin-related proteins, DRP2A and DRP2B, function coordinately in post-Golgi trafficking. Biochem. Biophys. Res. Commun. 2015, 456, 238–244. [Google Scholar] [CrossRef]
- Bednarek, S.Y.; Backues, S.K. Plant dynamin-related protein families DRP1 and DRP2 in plant development. Biochem. Soc. Trans. 2010, 38, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; An, C.; Liu, X.; Gao, H. Advances in molecular mechanisms of chloroplast division. J. Plant Physiol. 2016, 11, 1733–1744. [Google Scholar]
- Yoshida, Y.; Kuroiwa, H.; Misumi, O.; Nishida, K.; Yagisawa, F.; Fujiwara, T.; Nanamiya, H.; Kawamura, F.; Kuroiwa, T. Isolated chloroplast division machinery can actively constrict after stretching. Science 2006, 313, 1435–1438. [Google Scholar] [CrossRef]
- Yuan, B.; Liao, X.; Zheng, X.; Wu, L.; Zhao, H. Metabolism and role of Indoleacetic Acid in Plant cells. Bull. Biol. 2005, 40, 21–23. [Google Scholar]
- Zheng, Y.; Hu, W.; Jiang, A.; Bi, Y. Research of methyl jasmonate on the wounding defense response for fresh-cut sweet potatoes. Sci. Technol. Food Ind. 2012, 2, 368–372. [Google Scholar]
- Hannezo, E.; Scheele, C.L.G.J.; Moad, M.; Drogo, N.; Heer, R.; Sampogna, R.V.; Van Rheenen, J.; Simons, B.D. A Unifying Theory of Branching Morphogenesis. Cell 2017, 171, 242–255.e27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Guo, A.-Y. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.; Fu, L.; Yan, Y.; Tie, W.; Xia, Z.; Wang, W.; Peng, M.; Hu, W.; Zhang, J. Genome-wide characterization and expression profiling of HD-Zip gene family related to abiotic stress in cassava. PLoS ONE 2017, 12, e0173043. [Google Scholar] [CrossRef]
- Yao, Y.; Geng, M.T.; Wu, X.H.; Liu, J.; Li, R.M.; Hu, X.W.; Guo, J.C. Genome-wide identification, 3D modeling, expression and enzymatic activity analysis of cell wall invertase gene family from cassava (Manihot esculenta Crantz). Int. J. Mol. Sci. 2014, 15, 7313–7331. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Ali, M.; Luo, D.-X.; Khan, A.; Haq, S.U.; Gai, W.-X.; Zhang, H.-X.; Cheng, G.-X.; Muhammad, I.; Gong, Z.-H. Classification and Genome-Wide Analysis of Chitin-Binding Proteins Gene Family in Pepper (Capsicum annuum L.) and Transcriptional Regulation to Phytophthora capsici, Abiotic Stresses and Hormonal Applications. Int. J. Mol. Sci. 2018, 19, 2216. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, P.; Liu, X.; Guo, J.; Chen, Y.; Li, S.; Wang, C.; Huang, W.; Min, Y. Genome-Wide Analysis of Dynamin Gene Family in cassava (Manihot esculenta Crantz) and Transcriptional Regulation of Family Members ARC5 in Hormonal Treatments. Int. J. Mol. Sci. 2019, 20, 5094. https://doi.org/10.3390/ijms20205094
Cao P, Liu X, Guo J, Chen Y, Li S, Wang C, Huang W, Min Y. Genome-Wide Analysis of Dynamin Gene Family in cassava (Manihot esculenta Crantz) and Transcriptional Regulation of Family Members ARC5 in Hormonal Treatments. International Journal of Molecular Sciences. 2019; 20(20):5094. https://doi.org/10.3390/ijms20205094
Chicago/Turabian StyleCao, Peng, Xiaohan Liu, Jianchun Guo, Yinhua Chen, Shuangbao Li, Congcong Wang, Wu Huang, and Yi Min. 2019. "Genome-Wide Analysis of Dynamin Gene Family in cassava (Manihot esculenta Crantz) and Transcriptional Regulation of Family Members ARC5 in Hormonal Treatments" International Journal of Molecular Sciences 20, no. 20: 5094. https://doi.org/10.3390/ijms20205094



