Presence of Round Cells Proteins do not Interfere with Identification of Human Sperm Proteins from Frozen Semen Samples by LC-MS/MS
Abstract
:1. Introduction
2. Results
2.1. Semen Parameters
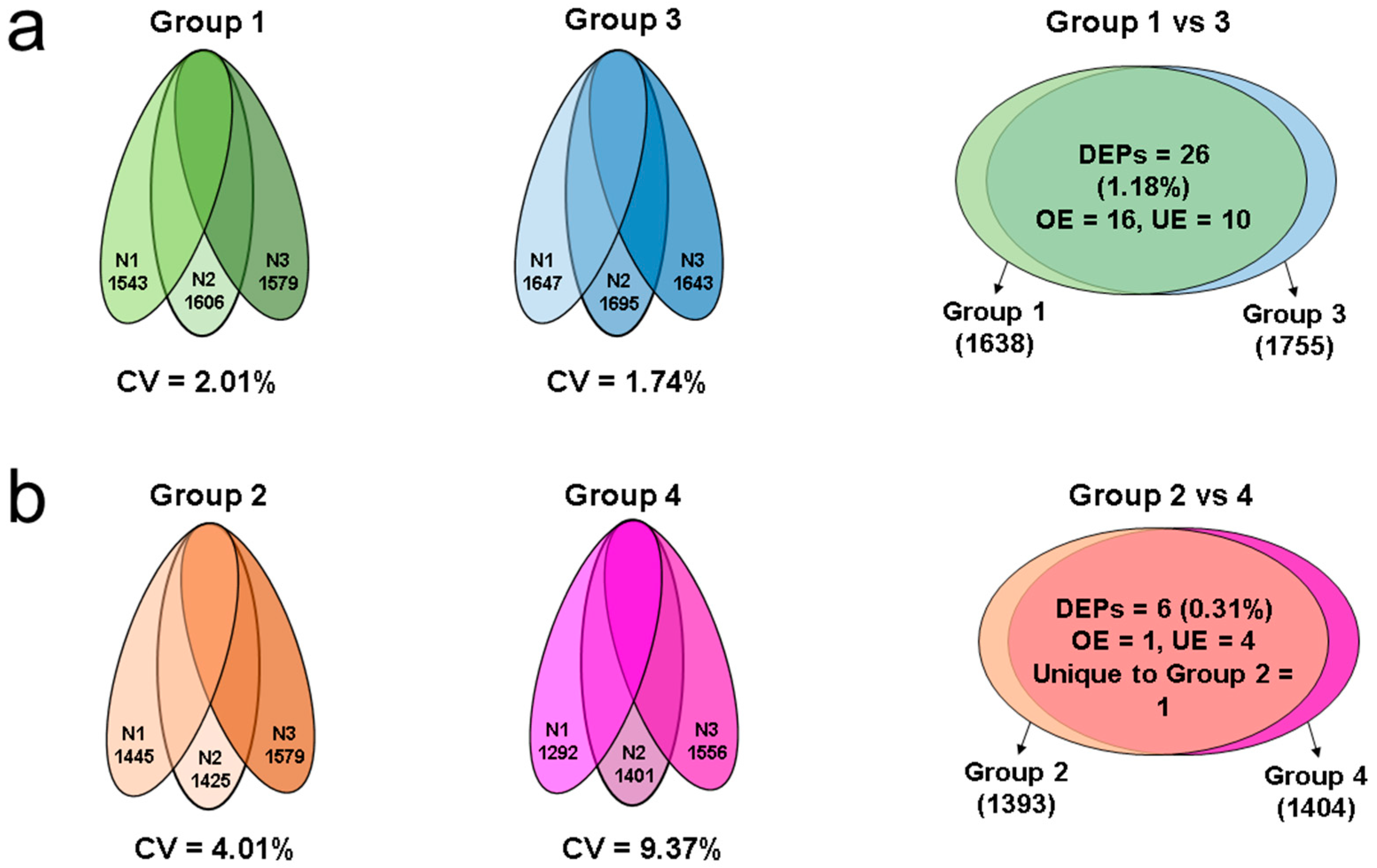
2.2. Proteomic Profile of Spermatozoa in Different Groups
2.3. Comparative Analysis of Human Sperm Protein and Leukocyte Protein
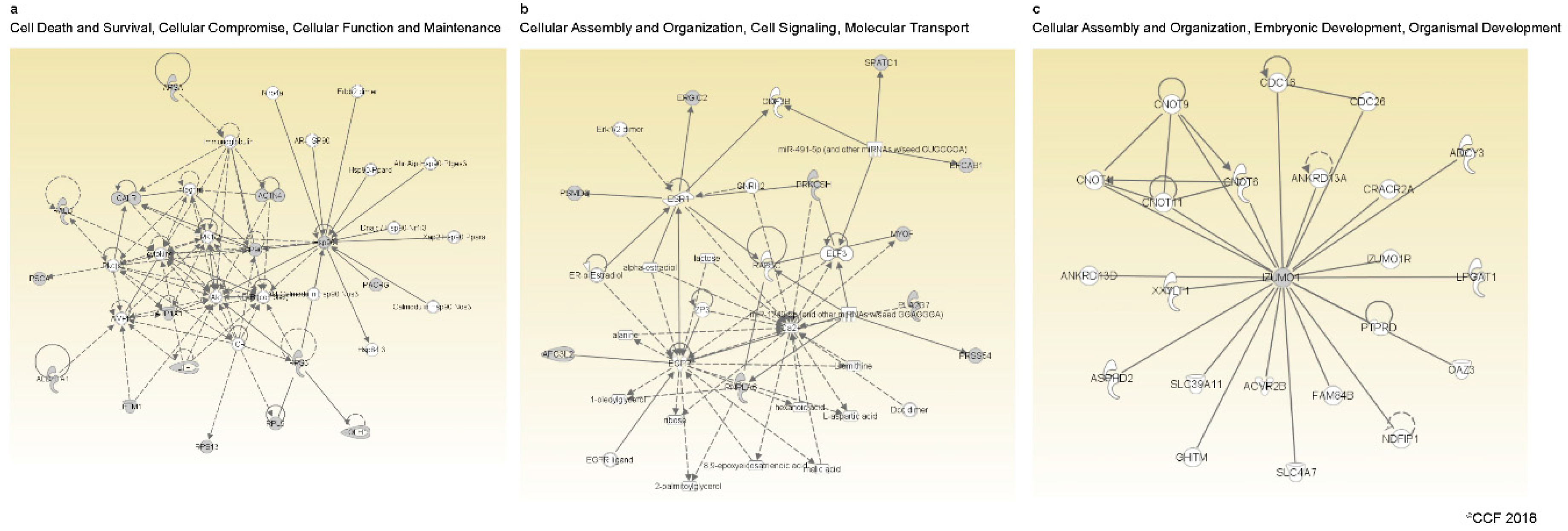
2.4. Molecular Pathways Regulated by DEPs in Spermatozoa
3. Discussion
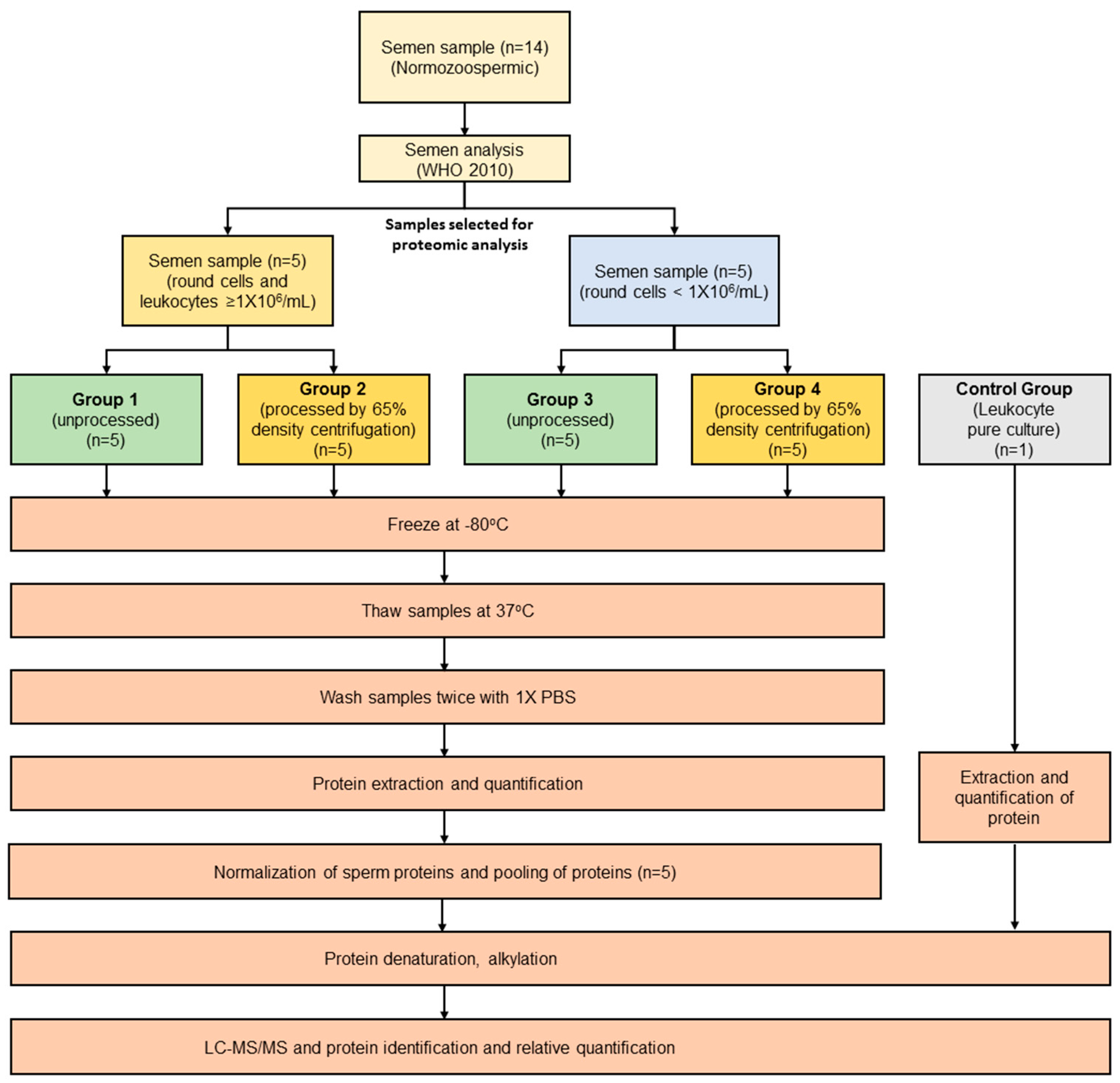
4. Materials and Methods
4.1. Semen Analysis
4.2. Inclusion and Exclusion Criteria
4.3. Processing of Semen Samples
4.4. Experimental Groups
4.5. Preparation of Samples for Proteomic Analysis
4.6. Quantitative Proteomic Analysis
4.7. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
4.8. Database Searching
4.9. Criteria for Protein Identification
4.10. Comparative Proteomics and Identification of Differentially Expressed Proteins
4.11. Bioinformatic Analysis of DEPs
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ganong, W.F.; Ganong, W. Review of Medical Physiology; Appleton & Lange: Norwalk, CT, USA, 1995. [Google Scholar]
- Samanta, L.; Parida, R.; Dias, T.R.; Agarwal, A. The enigmatic seminal plasma: A proteomics insight from ejaculation to fertilization. Reprod. Biol. Endocrinol. 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Johanisson, E.; Campana, A.; Luthi, R.; De Agostini, A. Evaluation ofround cells’ in semen analysis: A comparative study. Hum. Reprod. Update 2000, 6, 404–412. [Google Scholar] [CrossRef]
- Palermo, G.D.; Neri, Q.V.; Cozzubbo, T.; Cheung, S.; Pereira, N.; Rosenwaks, Z. Shedding Light on the Nature of Seminal Round Cells. PLOS ONE 2016, 11, e0151640. [Google Scholar] [CrossRef] [PubMed]
- De Bellabarba, G.A.; Tortolero, I.; Villarroel, V.; Molina, C.Z.; Bellabarba, C.; Velazquez, E. Nonsperm cells in human semen and their relationship with semen parameters. Arch. Androl. 2000, 45, 131–136. [Google Scholar]
- Majzoub, A.; Esteves, S.C.; Gosálvez, J.; Agarwal, A. Specialized sperm function tests in varicocele and the future of andrology laboratory. Asian J. Androl. 2016, 18, 205–212. [Google Scholar] [CrossRef]
- Agarwal, A.; Gupta, S.; Sharma, R. Leukocytospermia Quantitation (ENDTZ) Test. In Andrological Evaluation of Male Infertility: A Laboratory Guide; Agarwal, A., Gupta, S., Sharma, R., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 69–72. [Google Scholar]
- Shafer, M.-A.; Beck, A.; Blain, B.; Dole, P.; Irwin, C.E.; Sweet, R.; Schachter, J. Chlamydia trachomatis: Important relationships to race, contraception, lower genital tract infection, and Papanicolaou smear. J. Pediatr. 1984, 104, 141–146. [Google Scholar] [CrossRef]
- Saleh, R.A.; Agarwal, A.; Sharma, R.K.; Nelson, D.R.; Thomas, A.J., Jr. Effect of cigarette smoking on levels of seminal oxidative stress in infertile men: A prospective study. Fertil. Steril. 2002, 78, 491–499. [Google Scholar] [CrossRef]
- Maneesh, M.; Dutta, S.; Chakrabarti, A.; Vasudevan, D. Alcohol abuse-duration dependent decrease in plasma testosterone and antioxidants in males. Indian J. Physiol. Pharmacol. 2006, 50, 291–296. [Google Scholar]
- Agarwal, A.; Mulgund, A.; Alshahrani, S.; Assidi, M.; Abuzenadah, A.M.; Sharma, R.; Sabanegh, E. Reactive oxygen species and sperm DNA damage in infertile men presenting with low level leukocytospermia. Reprod. Biol. Endocrinol. 2014, 12. [Google Scholar] [CrossRef]
- Henkel, R.R.; Schill, W.B. Sperm preparation for ART. Reprod. Biol. Endocrinol. 2003, 1, 108. [Google Scholar] [CrossRef]
- Beydola, T.; Sharma, R.K.; Lee, W.; Agarwal, A. Sperm preparation and selection techniques. Male Infertil. Pract. 2013, 29, 244–251. [Google Scholar]
- Allamaneni, S.S.; Agarwal, A.; Rama, S.; Ranganathan, P.; Sharma, R.K. Comparative study on density gradients and swim-up preparation techniques utilizing neat and cryopreserved spermatozoa. Asian J. Androl. 2005, 7, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Panner Selvam, M.K.; Agarwal, A. Update on the proteomics of male infertility: A systematic review. Arab J. Urol. 2018, 16, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Amaral, A.; Castillo, J.; Ramalho-Santos, J.; Oliva, R. The combined human sperm proteome: Cellular pathways and implications for basic and clinical science. Hum. Reprod. Update 2014, 20, 40–62. [Google Scholar] [CrossRef]
- Wang, S.; Wang, W.; Xu, Y.; Tang, M.; Fang, J.; Sun, H.; Sun, Y.; Gu, M.; Liu, Z.; Zhang, Z. Proteomic characteristics of human sperm cryopreservation. Proteomics 2014, 14, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Amaral, A.; Paiva, C.; Attardo Parrinello, C.; Estanyol, J.M.; Ballescà, J.L.; Ramalho-Santos, J.; Oliva, R. Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J. Proteome Res. 2014, 13, 5670–5684. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Heredia, J.; Estanyol, J.M.; Ballescà, J.L.; Oliva, R. Proteomic identification of human sperm proteins. Proteomics 2006, 6, 4356–4369. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.M.; Xiang, Z.; Fu, Y.; Wu, H.L.; Zhu, W.B.; Fan, L.Q. Comparative Proteomics Reveal the Association between SPANX Proteins and Clinical Outcomes of Artificial Insemination with Donor Sperm. Sci. Rep. 2018, 8, 6850. [Google Scholar] [CrossRef]
- Intasqui, P.; Camargo, M.; Del Giudice, P.T.; Spaine, D.M.; Carvalho, V.M.; Cardozo, K.H.M.; Cedenho, A.P.; Bertolla, R.P. Unraveling the sperm proteome and post-genomic pathways associated with sperm nuclear DNA fragmentation. J. Assist. Reprod. Genet. 2013, 30, 1187–1202. [Google Scholar] [CrossRef] [Green Version]
- Bogle, O.; Kumar, K.; Attardo-Parrinello, C.; Lewis, S.; Estanyol, J.; Ballescà, J.; Oliva, R. Identification of protein changes in human spermatozoa throughout the cryopreservation process. Andrology 2017, 5, 10–22. [Google Scholar] [CrossRef]
- Intasqui, P.; Agarwal, A.; Sharma, R.; Samanta, L.; Bertolla, R. Towards the identification of reliable sperm biomarkers for male infertility: A sperm proteomic approach. Andrologia 2018, 50, e12919. [Google Scholar] [CrossRef]
- Samanta, L.; Agarwal, A.; Swain, N.; Sharma, R.; Gopalan, B.; Esteves, S.C.; Durairajanayagam, D.; Sabanegh, E. Proteomic Signatures of Sperm Mitochondria in Varicocele: Clinical Utility as Biomarkers of Varicocele Associated Infertility. J. Urol. 2018, 200, 414–422. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th ed.; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Dad, B.R.; Li-Jun, H. Posttranslational Modifications in Spermatozoa and Effects on Male Fertility and Sperm Viability. Omics J. Integr. Biol. 2017, 21, 245–256. [Google Scholar]
- Samanta, L.; Swain, N.; Ayaz, A.; Venugopal, V.; Agarwal, A. Post-Translational Modifications in sperm Proteome: The Chemistry of Proteome diversifications in the Pathophysiology of male factor infertility. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 1450–1465. [Google Scholar] [CrossRef] [PubMed]
- Vizel, R.; Hillman, P.; Ickowicz, D.; Breitbart, H. AKAP3 degradation in sperm capacitation is regulated by its tyrosine phosphorylation. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1912–1920. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Qi, H. Sperm-specific AKAP3 is a dual-specificity anchoring protein that interacts with both protein kinase a regulatory subunits via conserved N-terminal amphipathic peptides. Mol. Reprod. Dev. 2014, 81, 595–607. [Google Scholar] [CrossRef]
- Rahman, M.S.; Kwon, W.S.; Yoon, S.J.; Park, Y.J.; Ryu, B.Y.; Pang, M.G. A novel approach to assessing bisphenol-A hazards using an in vitro model system. BMC Genom. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Sharma, R.; Agarwal, A. Proteomic analysis of mature and immature ejaculated spermatozoa from fertile men. Asian J. Androl. 2016, 18, 735–746. [Google Scholar]
- Raverdeau, M.; Gely-Pernot, A.; Féret, B.; Dennefeld, C.; Benoit, G.; Davidson, I.; Chambon, P.; Mark, M.; Ghyselinck, N.B. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc. Natl. Acad. Sci. USA 2012, 109, 16582–16587. [Google Scholar] [CrossRef] [Green Version]
- Ikawa, M.; Tokuhiro, K.; Yamaguchi, R.; Benham, A.M.; Tamura, T.; Wada, I.; Satouh, Y.; Inoue, N.; Okabe, M. Calsperin is a testis-specific chaperone required for sperm fertility. J. Biol. Chem. 2011, 286, 5639–5646. [Google Scholar] [CrossRef]
- Redgrove, K.A.; Nixon, B.; Baker, M.A.; Hetherington, L.; Baker, G.; Liu, D.-Y.; Aitken, R.J. The Molecular Chaperone HSPA2 Plays a Key Role in Regulating the Expression of Sperm Surface Receptors That Mediate Sperm-Egg Recognition. PLoS ONE 2012, 7, e50851. [Google Scholar] [CrossRef] [PubMed]
- Weerachatyanukul, W.; Xu, H.; Anupriwan, A.; Carmona, E.; Wade, M.; Hermo, L.; da Silva, S.M.; Rippstein, P.; Sobhon, P.; Sretarugsa, P.; et al. Acquisition of Arylsulfatase A onto the Mouse Sperm Surface During Epididymal Transit1. Biol. Reprod. 2003, 69, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Widgren, E.E.; Richardson, R.T.; O’Rand, M.G. Characterization of an Eppin Protein Complex from Human Semen and Spermatozoa1. Biol. Reprod. 2007, 77, 476–484. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Agarwal, A.; Mohanty, G.; Du Plessis, S.S.; Gopalan, B.; Willard, B.; Yadav, S.P.; Sabanegh, E. Proteomic analysis of seminal fluid from men exhibiting oxidative stress. Reprod. Biol. Endocrinol. 2013, 11, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, N.; Hamada, D.; Kamikubo, H.; Hirata, K.; Kataoka, M.; Yamamoto, M.; Ikawa, M.; Okabe, M.; Hagihara, Y. Molecular dissection of IZUMO1, a sperm protein essential for sperm-egg fusion. Development 2013, 140, 3221–3229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohto, U.; Ishida, H.; Krayukhina, E.; Uchiyama, S.; Inoue, N.; Shimizu, T. Structure of IZUMO1–JUNO reveals sperm–oocyte recognition during mammalian fertilization. Nature 2016, 534, 566. [Google Scholar] [CrossRef] [PubMed]
- Newton, L.D. Na(+)/K(+)ATPase Regulates Sperm Capacitation Through a Mechanism Involving Kinases and Redistribution of Its Testis-Specific Isoform. Mol. Reprod. Dev. 2010, 77, 136–148. [Google Scholar] [CrossRef]
- Rajamanickam, G.D.; Kastelic, J.P.; Thundathil, J.C. Na/K-ATPase regulates bovine sperm capacitation through raft- and non-raft-mediated signaling mechanisms. Mol. Reprod. Dev. 2017, 84, 1168–1182. [Google Scholar] [CrossRef]
- Sutovsky, P. Sperm proteasome and fertilization. Reproduction 2011, 142, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Morales, P.; Kong, M.; Pizarro, E.; Pasten, C. Participation of the sperm proteasome in human fertilization. Hum. Reprod. 2003, 18, 1010–1017. [Google Scholar] [CrossRef] [Green Version]
- Panner Selvam, M.; Agarwal, A.; Sharma, R.; Samanta, L. Treatment of semen samples with α-chymotrypsin alters the expression pattern of sperm functional proteins—A pilot study. Andrology 2018, 6, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Ayaz, A.; Samanta, L.; Sharma, R.; Assidi, M.; Abuzenadah, A.M.; Sabanegh, E. Comparative proteomic network signatures in seminal plasma of infertile men as a function of reactive oxygen species. Clin. Proteom. 2015, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, A.; Agarwal, A.; Sharma, R.; Arafa, M.; Elbardisi, H.; Cui, Z. Impact of precise modulation of reactive oxygen species levels on spermatozoa proteins in infertile men. Clin. Proteom. 2015, 12, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, R.; Agarwal, A.; Mohanty, G.; Jesudasan, R.; Gopalan, B.; Willard, B.; Yadav, S.P.; Sabanegh, E. Functional proteomic analysis of seminal plasma proteins in men with various semen parameters. Reprod. Biol. Endocrinol. 2013, 11, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diz, A.P.; Truebano, M.; Skibinski, D.O. The consequences of sample pooling in proteomics: An empirical study. Electrophoresis 2009, 30, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
| Uniprot No. | Gene Name | Protein Name | Group 1 | Group 3 | NSAF Ratio | Expression | ||
|---|---|---|---|---|---|---|---|---|
| SC | Abun | SC | Abun | |||||
| P15289 | ARSA | Arylsulfatase A | 34.0 | M | 1.7 | VL | 0.05 | UE |
| P00352 | ALDH1A1 | Retinal dehydrogenase 1 | 26.3 | M | 2.7 | VL | 0.09 | UE |
| Q6PEW0 | PRSS54 | Inactive serine protease 54 | 18.0 | L | 4.7 | VL | 0.22 | UE |
| Q76KD6 | SPATC1 | Speriolin | 8.3 | L | 2.3 | VL | 0.26 | UE |
| Q9HAE3 | EFCAB1 | EF-hand calcium-binding domain-containing protein 1 | 9.3 | L | 3.3 | VL | 0.33 | UE |
| O43707 | ACTN4 | Alpha-actinin-4 | 44.7 | M | 23.3 | L | 0.47 | UE |
| P02788 | LTF | Lactotransferrin | 2486.7 | H | 1299.0 | H | 0.47 | UE |
| P14314 | PRKCSH | Glucosidase 2 subunit beta | 123.0 | H | 65.0 | M | 0.48 | UE |
| P27797 | CALR | Calreticulin | 271.7 | H | 175.0 | H | 0.57 | UE |
| P14625 | HSP90B1 | Endoplasmin | 987.0 | H | 660.3 | H | 0.60 | UE |
| P05023-4 | ATP1A1 | Isoform 4 of Sodium/potassium-transporting ATPase subunit alpha-1 | 98.7 | H | 181.3 | H | 1.61 | OE |
| O95202 | LETM1 | Mitochondrial proton/calcium exchanger protein | 29.0 | M | 65.7 | M | 2.04 | OE |
| O43242 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3 | 33.7 | M | 78.3 | M | 2.04 | OE |
| P23396 | RPS3 | 40S ribosomal protein S3 | 24.7 | M | 60.0 | M | 2.20 | OE |
| P11234 | RALB | Ras-related protein Ral-B | 9.0 | L | 22.0 | M | 2.25 | OE |
| Q9NZM1-3 | MYOF | Isoform 3 of Myoferlin | 21.3 | M | 55.3 | M | 2.27 | OE |
| Q9Y4W6 | AFG3L2 | AFG3-like protein 2 | 15.3 | L | 40.7 | M | 2.38 | OE |
| Q96RQ1 | ERGIC2 | Endoplasmic reticulum-Golgi intermediate compartment protein 2 | 8.3 | L | 23.0 | M | 2.52 | OE |
| Q8IYV9 | IZUMO1 | Izumo sperm-egg fusion protein 1 | 9.0 | L | 28.7 | M | 3.00 | OE |
| Q04609 | FOLH1 | Glutamate carboxypeptidase 2 | 27.0 | M | 96.3 | H | 3.21 | OE |
| Q8IY17-4 | PNPLA6 | Isoform 4 of Neuropathy target esterase | 3.7 | VL | 14.0 | L | 3.36 | OE |
| P62277 | RPS13 | 40S ribosomal protein S13 | 5.3 | VL | 20.7 | M | 3.42 | OE |
| P46777 | RPL5 | 60S ribosomal protein L5 | 5.7 | VL | 24.7 | M | 3.93 | OE |
| Q13093 | PLA2G7 | Platelet-activating factor acetylhydrolase | 5.3 | VL | 22.7 | M | 4.02 | OE |
| O43653 | PSCA | Prostate stem cell antigen | 1.7 | VL | 14.3 | L | 7.22 | OE |
| Q96M98-2 | PACRG | Isoform 2 of Parkin coregulated gene protein | 1.3 | VL | 21.0 | M | 13.04 | OE |
| Uniprot No. | Gene Name | Protein Name | Group 2 | Group 4 | NSAF Ratio | Expression | ||
|---|---|---|---|---|---|---|---|---|
| SC | Abun | SC | Abun | |||||
| O75969 | AKAP3 | A-kinase anchor protein 3 | 146.3 | H | 81.7 | H | 0.56 | UE |
| O96005 | CLPTM1 | Cleft lip and palate transmembrane protein 1 | 41.3 | M | 19.0 | L | 0.45 | UE |
| Q9Y619 | SLC25A15 | Mitochondrial ornithine transporter 1 | 26.0 | M | 12.0 | L | 0.47 | UE |
| Q8TF71 | SLC16A10 | Monocarboxylate transporter 10 | 2.0 | VL | 0.0 | - | 0.00 | Unique to Group 2 |
| Q96ND0 | FAM210A | Protein FAM210A | 5.0 | VL | 0.7 | VL | 0.14 | OE |
| P12074 | COX6A1 | Cytochrome c oxidase subunit 6A1, mitochondrial | 10.0 | L | 21.0 | M | 2.15 | OE |
| Uniprot No. | Gene Name | Protein Name | Group 1 | Group 3 | Control Group (CG) | NSAF Ratio CG/ Group1 | Expression in CG | |||
|---|---|---|---|---|---|---|---|---|---|---|
| SC | Abun | SC | Abun | SC | Abun | |||||
| P00352 | ALDH1A1 | Retinal dehydrogenase 1 | 26.3 | M | 2.7 | VL | 0.0 | - | 0.00 | Absent in CG |
| Q6PEW0 | PRSS54 | Inactive serine protease 54 | 18.0 | L | 4.7 | VL | 0.0 | - | 0.00 | Absent in CG |
| Q76KD6 | SPATC1 | Speriolin | 8.3 | L | 2.3 | VL | 0.0 | - | 0.00 | Absent in CG |
| Q9HAE3 | EFCAB1 | EF-hand calcium-binding domain-containing protein 1 | 9.3 | L | 3.3 | VL | 0.0 | - | 0.00 | Absent in CG |
| O43242 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3 | 33.7 | M | 78.3 | M | 0.0 | - | 0.00 | Absent in CG |
| P23396 | RPS3 | 40S ribosomal protein S3 | 24.7 | M | 60.0 | M | 0.0 | - | 0.00 | Absent in CG |
| Q9NZM1-3 | MYOF | Isoform 3 of Myoferlin | 21.3 | M | 55.3 | M | 0.0 | - | 0.00 | Absent in CG |
| Q96RQ1 | ERGIC2 | Endoplasmic reticulum-Golgi intermediate compartment protein 2 | 8.3 | L | 23.0 | M | 0.0 | - | 0.00 | Absent in CG |
| Q8IYV9 | IZUMO1 | Izumo sperm-egg fusion protein 1 | 9.0 | L | 28.7 | M | 0.0 | - | 0.00 | Absent in CG |
| Q04609 | FOLH1 | Glutamate carboxypeptidase 2 | 27.0 | M | 96.3 | H | 0.0 | - | 0.00 | Absent in CG |
| Q8IY17-4 | PNPLA6 | Isoform 4 of Neuropathy target esterase | 3.7 | VL | 14.0 | L | 0.0 | - | 0.00 | Absent in CG |
| P62277 | RPS13 | 40S ribosomal protein S13 | 5.3 | VL | 20.7 | M | 0.0 | - | 0.00 | Absent in CG |
| Q13093 | PLA2G7 | Platelet-activating factor acetylhydrolase | 5.3 | VL | 22.7 | M | 0.0 | - | 0.00 | Absent in CG |
| O43653 | PSCA | Prostate stem cell antigen | 1.7 | VL | 14.3 | L | 0.0 | - | 0.00 | Absent in CG |
| Q96M98-2 | PACRG | Isoform 2 of Parkin coregulated gene protein | 1.3 | VL | 21.0 | M | 0.0 | - | 0.00 | Absent in CG |
| P46777 | RPL5 | 60S ribosomal protein L5 | 5.7 | VL | 24.7 | M | 1.3 | M | 0.36 | UE |
| P15289 | ARSA | Arylsulfatase A | 34.0 | M | 1.7 | VL | 4.7 | VL | 0.20 | UE |
| P02788 | LTF | Lactotransferrin | 2486.7 | H | 1299.0 | H | 424.3 | H | 0.24 | UE |
| P14314 | PRKCSH | Glucosidase 2 subunit beta | 123.0 | H | 65.0 | M | 33.7 | M | 0.39 | UE |
| P27797 | CALR | Calreticulin | 271.7 | H | 175.0 | H | 123.0 | H | 0.65 | UE |
| P14625 | HSP90B1 | Endoplasmin | 987.0 | H | 660.3 | H | 73.3 | M | 0.11 | UE |
| P05023-4 | ATP1A1 | Isoform 4 of Sodium/potassium-transporting ATPase subunit alpha-1 | 98.7 | H | 181.3 | H | 27.3 | M | 0.39 | UE |
| O95202 | LETM1 | Mitochondrial proton/calcium exchanger protein | 29.0 | M | 65.7 | M | 9.0 | L | 0.46 | UE |
| Q9Y4W6 | AFG3L2 | AFG3-like protein 2 | 15.3 | L | 40.7 | M | 0.7 | VL | 0.06 | UE |
| P11234 | RALB | Ras-related protein Ral-B | 9.0 | L | 22.0 | M | 62.0 | M | 10.12 | OE |
| O43707 | ACTN4 | Alpha-actinin-4 | 44.7 | M | 23.3 | L | 52.3 | M | 1.73 | OE |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panner Selvam, M.K.; Agarwal, A.; Dias, T.R.; Martins, A.D.; Samanta, L. Presence of Round Cells Proteins do not Interfere with Identification of Human Sperm Proteins from Frozen Semen Samples by LC-MS/MS. Int. J. Mol. Sci. 2019, 20, 314. https://doi.org/10.3390/ijms20020314
Panner Selvam MK, Agarwal A, Dias TR, Martins AD, Samanta L. Presence of Round Cells Proteins do not Interfere with Identification of Human Sperm Proteins from Frozen Semen Samples by LC-MS/MS. International Journal of Molecular Sciences. 2019; 20(2):314. https://doi.org/10.3390/ijms20020314
Chicago/Turabian StylePanner Selvam, Manesh Kumar, Ashok Agarwal, Tânia R. Dias, Ana D. Martins, and Luna Samanta. 2019. "Presence of Round Cells Proteins do not Interfere with Identification of Human Sperm Proteins from Frozen Semen Samples by LC-MS/MS" International Journal of Molecular Sciences 20, no. 2: 314. https://doi.org/10.3390/ijms20020314
APA StylePanner Selvam, M. K., Agarwal, A., Dias, T. R., Martins, A. D., & Samanta, L. (2019). Presence of Round Cells Proteins do not Interfere with Identification of Human Sperm Proteins from Frozen Semen Samples by LC-MS/MS. International Journal of Molecular Sciences, 20(2), 314. https://doi.org/10.3390/ijms20020314







