Molecular Mechanisms of Leucine Zipper EF-Hand Containing Transmembrane Protein-1 Function in Health and Disease
Abstract
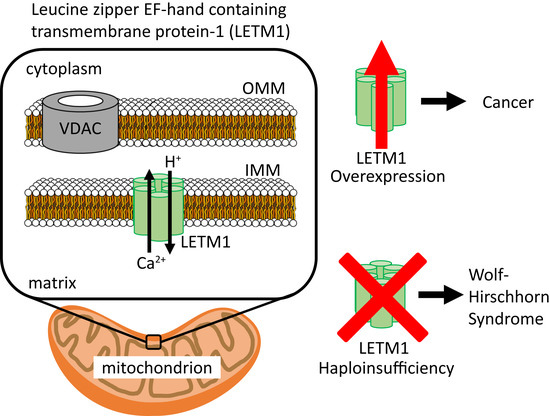
:1. Introduction
2. LETM1 Protein Family Domain Architecture and Homology
2.1. The LETM1 Protein Family
2.2. LETM1 Domain Architecture and Homology
3. The Role of LETM1 in Mitochondrial Physiology
3.1. LETM1 in K+/H+ Exchange
3.2. LETM1 in Ca2+/H+ Exchange
3.3. LETM1 Protein Structure and Assembly
4. Wolf-Hirschhorn Syndrome (WHS) and other LETM1-Associated Pathophysiologies
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Brini, M.; Carafoli, E.; Cali, T. The plasma membrane calcium pumps: Focus on the role in (neuro)pathology. Biochem. Biophys. Res. Commun. 2017, 483, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, M.; Nishikawa, T.; Siddiqui, N.; Chung, S.; Ikura, M.; Stathopulos, P.B. From Stores to Sinks: Structural Mechanisms of Cytosolic Calcium Regulation. Adv. Exp. Med. Biol. 2017, 981, 215–251. [Google Scholar] [PubMed]
- Krebs, J.; Agellon, L.B.; Michalak, M. Ca2+ homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochem. Biophys. Res. Commun. 2015, 460, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Putney, J.W.; Steinckwich-Besancon, N.; Numaga-Tomita, T.; Davis, F.M.; Desai, P.N.; D’Agostin, D.M.; Wu, S.; Bird, G.S. The functions of store-operated calcium channels. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Lanner, J.T.; Georgiou, D.K.; Joshi, A.D.; Hamilton, S.L. Ryanodine receptors: Structure, expression, molecular details, and function in calcium release. Cold Spring Harb. Perspect. Biol. 2010, 2, a003996. [Google Scholar] [CrossRef]
- Prole, D.L.; Taylor, C.W. Inositol 1,4,5-trisphosphate receptors and their protein partners as signalling hubs. J. Physiol. 2016, 594, 2849–2866. [Google Scholar] [CrossRef] [Green Version]
- Santulli, G.; Lewis, D.; des Georges, A.; Marks, A.R.; Frank, J. Ryanodine Receptor Structure and Function in Health and Disease. Subcell. Biochem. 2018, 87, 329–352. [Google Scholar]
- Santulli, G.; Nakashima, R.; Yuan, Q.; Marks, A.R. Intracellular calcium release channels: An update. J. Physiol. 2017, 595, 3041–3051. [Google Scholar] [CrossRef]
- Van Petegem, F. Ryanodine receptors: Allosteric ion channel giants. J. Mol. Biol. 2015, 427, 31–53. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Ikura, M. Store operated calcium entry: From concept to structural mechanisms. Cell Calcium 2017, 63, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Lunz, V.; Romanin, C.; Frischauf, I. STIM1 activation of Orai1. Cell Calcium 2019, 77, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Novello, M.J.; Zhu, J.; Feng, Q.; Ikura, M.; Stathopulos, P.B. Structural elements of stromal interaction molecule function. Cell Calcium 2018, 73, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Zhang, M.; Stathopulos, P.B. The 2beta Splice Variation Alters the Structure and Function of the Stromal Interaction Molecule Coiled-Coil Domains. Int. J. Mol. Sci. 2018, 19, 3316. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lu, X.; Feng, Q.; Stathopulos, P.B. A charge-sensing region in the stromal interaction molecule 1 luminal domain confers stabilization-mediated inhibition of SOCE in response to S-nitrosylation. J. Biol. Chem. 2018, 293, 8900–8911. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Ikura, M. Does stromal interaction molecule-1 have five senses? Cell Calcium 2019, 77, 79–80. [Google Scholar] [CrossRef]
- Manjarres, I.M.; Rodriguez-Garcia, A.; Alonso, M.T.; Garcia-Sancho, J. The sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) is the third element in capacitative calcium entry. Cell Calcium 2010, 47, 412–418. [Google Scholar] [CrossRef]
- Gambetti, P.; Erulkar, S.E.; Somlyo, A.P.; Gonatas, N.K. Calcium-containing structures in vertebrate glial cells. Ultrastructural and microprobe analysis. J. Cell Biol. 1975, 64, 322–330. [Google Scholar] [CrossRef] [Green Version]
- Henkart, M.P.; Reese, T.S.; Brinley, F.J., Jr. Endoplasmic reticulum sequesters calcium in the squid giant axon. Science 1978, 202, 1300–1303. [Google Scholar] [CrossRef]
- Howell, S.L.; Montague, W.; Tyhurst, M. Calcium distribution in islets of Langerhans: A study of calcium concentrations and of calcium accumulation in B cell organelles. J. Cell Sci. 1975, 19, 395–409. [Google Scholar]
- Kendrick, N.C.; Blaustein, M.P.; Fried, R.C.; Ratzlaff, R.W. ATP-dependent calcium storage in presynaptic nerve terminals. Nature 1977, 265, 246–248. [Google Scholar] [CrossRef]
- Somlyo, A.P. Cell physiology: Cellular site of calcium regulation. Nature 1984, 309, 516–517. [Google Scholar] [CrossRef] [PubMed]
- Somlyo, A.P.; Bond, M.; Somlyo, A.V. Calcium content of mitochondria and endoplasmic reticulum in liver frozen rapidly in vivo. Nature 1985, 314, 622–625. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.P.; Masoro, E.J.; DeMartinis, F.D. Imidazole and sequestration of calcium ions by sarcoplasmic reticulum. Nature 1967, 216, 822–824. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Streb, H.; Irvine, R.F.; Berridge, M.J.; Schulz, I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 1983, 306, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Poburko, D.; Demaurex, N. Regulation of the mitochondrial proton gradient by cytosolic Ca2+ signals. Pflugers Arch. 2012, 464, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Drago, I.; Pizzo, P.; Pozzan, T. After half a century mitochondrial calcium in- and efflux machineries reveal themselves. EMBO J. 2011, 30, 4119–4125. [Google Scholar] [CrossRef] [Green Version]
- Foskett, J.K.; Philipson, B. The mitochondrial Ca2+ uniporter complex. J. Mol. Cell. Cardiol. 2015, 78, 3–8. [Google Scholar] [CrossRef]
- Rottenberg, H.; Scarpa, A. Calcium uptake and membrane potential in mitochondria. Biochemistry 1974, 13, 4811–4817. [Google Scholar] [CrossRef]
- Mitchell, P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. 1966. Biochim. Biophys. Acta 2011, 1807, 1507–1538. [Google Scholar] [CrossRef]
- Balaban, R.S. The role of Ca2+ signaling in the coordination of mitochondrial ATP production with cardiac work. Biochim. Biophys. Acta 2009, 1787, 1334–1341. [Google Scholar] [CrossRef] [PubMed]
- Denton, R.M. Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 2009, 1787, 1309–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutter, G.A.; Rizzuto, R. Regulation of mitochondrial metabolism by ER Ca2+ release: An intimate connection. Trends Biochem. Sci. 2000, 25, 215–221. [Google Scholar] [CrossRef]
- Tarasov, A.I.; Griffiths, E.J.; Rutter, G.A. Regulation of ATP production by mitochondrial Ca2+. Cell Calcium 2012, 52, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Pathak, T.; Trebak, M. Mitochondrial Ca2+ signaling. Pharmacol. Ther. 2018, 192, 112–123. [Google Scholar] [CrossRef]
- Bhosale, G.; Sharpe, J.A.; Koh, A.; Kouli, A.; Szabadkai, G.; Duchen, M.R. Pathological consequences of MICU1 mutations on mitochondrial calcium signalling and bioenergetics. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1009–1017. [Google Scholar] [CrossRef]
- Chaudhuri, D.; Artiga, D.J.; Abiria, S.A.; Clapham, D.E. Mitochondrial calcium uniporter regulator 1 (MCUR1) regulates the calcium threshold for the mitochondrial permeability transition. Proc. Natl. Acad. Sci. USA 2016, 113, E1872–E1880. [Google Scholar] [CrossRef]
- Alavian, K.N.; Beutner, G.; Lazrove, E.; Sacchetti, S.; Park, H.A.; Licznerski, P.; Li, H.; Nabili, P.; Hockensmith, K.; Graham, M.; et al. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. USA 2014, 111, 10580–10585. [Google Scholar] [CrossRef] [Green Version]
- Bonora, M.; Bononi, A.; De Marchi, E.; Giorgi, C.; Lebiedzinska, M.; Marchi, S.; Patergnani, S.; Rimessi, A.; Suski, J.M.; Wojtala, A.; et al. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell Cycle 2013, 12, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, V.; von Stockum, S.; Antoniel, M.; Fabbro, A.; Fogolari, F.; Forte, M.; Glick, G.D.; Petronilli, V.; Zoratti, M.; Szabo, I.; et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. USA 2013, 110, 5887–5892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halestrap, A. Biochemistry: A pore way to die. Nature 2005, 434, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Shanmughapriya, S.; Rajan, S.; Hoffman, N.E.; Higgins, A.M.; Tomar, D.; Nemani, N.; Hines, K.J.; Smith, D.J.; Eguchi, A.; Vallem, S.; et al. SPG7 Is an Essential and Conserved Component of the Mitochondrial Permeability Transition Pore. Mol. Cell 2015, 60, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Kinnally, K.W.; Peixoto, P.M.; Ryu, S.Y.; Dejean, L.M. Is mPTP the gatekeeper for necrosis, apoptosis, or both? Biochim. Biophys. Acta 2011, 1813, 616–622. [Google Scholar] [CrossRef] [Green Version]
- Nicotra, A.; Parvez, S. Apoptotic molecules and MPTP-induced cell death. Neurotoxicol. Teratol. 2002, 24, 599–605. [Google Scholar] [CrossRef]
- Baughman, J.M.; Perocchi, F.; Girgis, H.S.; Plovanich, M.; Belcher-Timme, C.A.; Sancak, Y.; Bao, X.R.; Strittmatter, L.; Goldberger, O.; Bogorad, R.L.; et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 2011, 476, 341–345. [Google Scholar] [CrossRef] [Green Version]
- De Stefani, D.; Raffaello, A.; Teardo, E.; Szabo, I.; Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 2011, 476, 336–340. [Google Scholar] [CrossRef] [Green Version]
- Kirichok, Y.; Krapivinsky, G.; Clapham, D.E. The mitochondrial calcium uniporter is a highly selective ion channel. Nature 2004, 427, 360–364. [Google Scholar] [CrossRef]
- Hoffman, N.E.; Chandramoorthy, H.C.; Shanmughapriya, S.; Zhang, X.Q.; Vallem, S.; Doonan, P.J.; Malliankaraman, K.; Guo, S.; Rajan, S.; Elrod, J.W.; et al. SLC25A23 augments mitochondrial Ca2+ uptake, interacts with MCU, and induces oxidative stress-mediated cell death. Mol. Biol. Cell 2014, 25, 936–947. [Google Scholar] [CrossRef]
- Perocchi, F.; Gohil, V.M.; Girgis, H.S.; Bao, X.R.; McCombs, J.E.; Palmer, A.E.; Mootha, V.K. MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 2010, 467, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Raffaello, A.; De Stefani, D.; Sabbadin, D.; Teardo, E.; Merli, G.; Picard, A.; Checchetto, V.; Moro, S.; Szabo, I.; Rizzuto, R. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013, 32, 2362–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancak, Y.; Markhard, A.L.; Kitami, T.; Kovacs-Bogdan, E.; Kamer, K.J.; Udeshi, N.D.; Carr, S.A.; Chaudhuri, D.; Clapham, D.E.; Li, A.A.; et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 2013, 342, 1379–1382. [Google Scholar] [CrossRef]
- Dong, Z.; Shanmughapriya, S.; Tomar, D.; Siddiqui, N.; Lynch, S.; Nemani, N.; Breves, S.L.; Zhang, X.; Tripathi, A.; Palaniappan, P.; et al. Mitochondrial Ca2+ Uniporter Is a Mitochondrial Luminal Redox Sensor that Augments MCU Channel Activity. Mol. Cell 2017, 65, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Shanmughapriya, S.; Mok, M.C.Y.; Dong, Z.; Tomar, D.; Carvalho, E.; Rajan, S.; Junop, M.S.; Madesh, M.; Stathopulos, P.B. Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations. Cell Chem. Biol. 2016, 23, 1157–1169. [Google Scholar] [CrossRef] [Green Version]
- Boyman, L.; Williams, G.S.; Khananshvili, D.; Sekler, I.; Lederer, W.J. NCLX: The mitochondrial sodium calcium exchanger. J. Mol. Cell. Cardiol. 2013, 59, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Palty, R.; Silverman, W.F.; Hershfinkel, M.; Caporale, T.; Sensi, S.L.; Parnis, J.; Nolte, C.; Fishman, D.; Shoshan-Barmatz, V.; Herrmann, S.; et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. USA 2010, 107, 436–441. [Google Scholar] [CrossRef]
- Doonan, P.J.; Chandramoorthy, H.C.; Hoffman, N.E.; Zhang, X.; Cardenas, C.; Shanmughapriya, S.; Rajan, S.; Vallem, S.; Chen, X.; Foskett, J.K.; et al. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. FASEB J. 2014, 28, 4936–4949. [Google Scholar] [CrossRef]
- Jiang, D.; Zhao, L.; Clish, C.B.; Clapham, D.E. Letm1, the mitochondrial Ca2+/H+ antiporter, is essential for normal glucose metabolism and alters brain function in Wolf-Hirschhorn syndrome. Proc. Natl. Acad. Sci. USA 2013, 110, E2249–E2254. [Google Scholar] [CrossRef]
- Jiang, D.; Zhao, L.; Clapham, D.E. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science 2009, 326, 144–147. [Google Scholar] [CrossRef]
- Shao, J.; Fu, Z.; Ji, Y.; Guan, X.; Guo, S.; Ding, Z.; Yang, X.; Cong, Y.; Shen, Y. Leucine zipper-EF-hand containing transmembrane protein 1 (LETM1) forms a Ca2+/H+ antiporter. Sci. Rep. 2016, 6, 34174. [Google Scholar] [CrossRef] [PubMed]
- Tamai, S.; Iida, H.; Yokota, S.; Sayano, T.; Kiguchiya, S.; Ishihara, N.; Hayashi, J.; Mihara, K.; Oka, T. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J. Cell Sci. 2008, 121 Pt 15, 2588–2600. [Google Scholar] [CrossRef] [Green Version]
- Tsai, M.F.; Jiang, D.; Zhao, L.; Clapham, D.; Miller, C. Functional reconstitution of the mitochondrial Ca2+/H+ antiporter Letm1. J. Gen. Physiol. 2014, 143, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Waldeck-Weiermair, M.; Jean-Quartier, C.; Rost, R.; Khan, M.J.; Vishnu, N.; Bondarenko, A.I.; Imamura, H.; Malli, R.; Graier, W.F. Leucine zipper EF hand-containing transmembrane protein 1 (Letm1) and uncoupling proteins 2 and 3 (UCP2/3) contribute to two distinct mitochondrial Ca2+ uptake pathways. J. Biol. Chem. 2011, 286, 28444–28455. [Google Scholar] [CrossRef] [PubMed]
- Endele, S.; Fuhry, M.; Pak, S.J.; Zabel, B.U.; Winterpacht, A. LETM1, a novel gene encoding a putative EF-hand Ca2+-binding protein, flanks the Wolf-Hirschhorn syndrome (WHS) critical region and is deleted in most WHS patients. Genomics 1999, 60, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Rauch, A.; Schellmoser, S.; Kraus, C.; Dorr, H.G.; Trautmann, U.; Altherr, M.R.; Pfeiffer, R.A.; Reis, A. First known microdeletion within the Wolf-Hirschhorn syndrome critical region refines genotype-phenotype correlation. Am. J. Med. Genet. 2001, 99, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Stec, I.; van Ommen, G.J.; den Dunnen, J.T. WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics 2001, 76, 5–8. [Google Scholar] [CrossRef]
- Hashimi, H.; McDonald, L.; Stribrna, E.; Lukes, J. Trypanosome Letm1 protein is essential for mitochondrial potassium homeostasis. J. Biol. Chem. 2013, 288, 26914–26925. [Google Scholar] [CrossRef] [PubMed]
- McQuibban, A.G.; Joza, N.; Megighian, A.; Scorzeto, M.; Zanini, D.; Reipert, S.; Richter, C.; Schweyen, R.J.; Nowikovsky, K. A Drosophila mutant of LETM1, a candidate gene for seizures in Wolf-Hirschhorn syndrome. Hum. Mol. Genet. 2010, 19, 987–1000. [Google Scholar] [CrossRef]
- Nowikovsky, K.; Froschauer, E.M.; Zsurka, G.; Samaj, J.; Reipert, S.; Kolisek, M.; Wiesenberger, G.; Schweyen, R.J. The LETM1/YOL027 gene family encodes a factor of the mitochondrial K+ homeostasis with a potential role in the Wolf-Hirschhorn syndrome. J. Biol. Chem. 2004, 279, 30307–30315. [Google Scholar] [CrossRef]
- Chazin, W.J. Relating form and function of EF-hand calcium binding proteins. Acc. Chem. Res. 2011, 44, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Kretsinger, R.H. Structural and functional diversity of EF-hand proteins: Evolutionary perspectives. Protein Sci. 2017, 26, 1898–1920. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef]
- Ikura, M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem. Sci. 1996, 21, 14–17. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Zheng, L.; Li, G.Y.; Plevin, M.J.; Ikura, M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell 2008, 135, 110–122. [Google Scholar] [CrossRef]
- Stathopulos, P.B.; Ikura, M. Structure and Function of Endoplasmic Reticulum STIM Calcium Sensors. In Current Topics in Membranes; Academic Press: Cambridge, MA, USA, 2013; Volume 71, pp. 59–93. [Google Scholar]
- Dimmer, K.S.; Navoni, F.; Casarin, A.; Trevisson, E.; Endele, S.; Winterpacht, A.; Salviati, L.; Scorrano, L. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum. Mol. Genet. 2008, 17, 201–214. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef] [Green Version]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef]
- Huang, X.Q.; Miller, W. A Time-Efficient, Linear-Space Local Similarity Algorithm. Adv. Appl. Math. 1991, 12, 337–357. [Google Scholar] [CrossRef]
- Baeza, J.; Smallegan, M.J.; Denu, J.M. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem. Biol. 2015, 10, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Shan, C.; Kang, H.B.; Elf, S.; Xie, J.; Tucker, M.; Gu, T.L.; Aguiar, M.; Lonning, S.; Chen, H.; et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol. Cell 2014, 53, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Webster, B.R.; Li, J.H.; Sack, M.N. Identification of a molecular component of the mitochondrial acetyltransferase programme: A novel role for GCN5L1. Biochem. J. 2012, 443, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.R.; Payne, R.M. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013, 288, 29036–29045. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [PubMed] [Green Version]
- Hasegawa, A.; van der Bliek, A.M. Inverse correlation between expression of the Wolfs Hirschhorn candidate gene Letm1 and mitochondrial volume in C. elegans and in mammalian cells. Hum. Mol. Genet. 2007, 16, 2061–2071. [Google Scholar] [CrossRef] [PubMed]
- Austin, S.; Tavakoli, M.; Pfeiffer, C.; Seifert, J.; Mattarei, A.; De Stefani, D.; Zoratti, M.; Nowikovsky, K. LETM1-Mediated K+ and Na+ Homeostasis Regulates Mitochondrial Ca2+ Efflux. Front. Physiol. 2017, 8, 839. [Google Scholar] [CrossRef] [PubMed]
- Nowikovsky, K.; Reipert, S.; Devenish, R.J.; Schweyen, R.J. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007, 14, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Garlid, K.D.; Paucek, P. Mitochondrial potassium transport: The K+ cycle. Biochim. Biophys. Acta 2003, 1606, 23–41. [Google Scholar] [CrossRef]
- Argese, E.; Bettiol, C.; Marchetto, D.; De Vettori, S.; Zambon, A.; Miana, P.; Ghetti, P.F. Study on the toxicity of phenolic and phenoxy herbicides using the submitochondrial particle assay. Toxicol. In Vitro 2005, 19, 1035–1043. [Google Scholar] [CrossRef]
- Froschauer, E.; Nowikovsky, K.; Schweyen, R.J. Electroneutral K+/H+ exchange in mitochondrial membrane vesicles involves Yol027/Letm1 proteins. Biochim. Biophys. Acta 2005, 1711, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Gwack, Y.; Prakriya, M.; Srikanth, S.; Puppel, S.H.; Tanasa, B.; Hogan, P.G.; Lewis, R.S.; Daly, M.; Rao, A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 2006, 441, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Vig, M.; Peinelt, C.; Beck, A.; Koomoa, D.L.; Rabah, D.; Koblan-Huberson, M.; Kraft, S.; Turner, H.; Fleig, A.; Penner, R.; et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 2006, 312, 1220–1223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Yeromin, A.V.; Zhang, X.H.; Yu, Y.; Safrina, O.; Penna, A.; Roos, J.; Stauderman, K.A.; Cahalan, M.D. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA 2006, 103, 9357–9362. [Google Scholar] [CrossRef]
- Liou, J.; Kim, M.L.; Heo, W.D.; Jones, J.T.; Myers, J.W.; Ferrell, J.E., Jr.; Meyer, T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005, 15, 1235–1241. [Google Scholar] [CrossRef] [PubMed]
- Roos, J.; DiGregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Pittman, J.K.; Shigaki, T.; Marshall, J.L.; Morris, J.L.; Cheng, N.H.; Hirschi, K.D. Functional and regulatory analysis of the Arabidopsis thaliana CAX2 cation transporter. Plant Mol. Biol. 2004, 56, 959–971. [Google Scholar] [CrossRef]
- Socha, A.L.; Guerinot, M.L. Mn-euvering manganese: The role of transporter gene family members in manganese uptake and mobilization in plants. Front. Plant Sci. 2014, 5, 106. [Google Scholar] [CrossRef]
- Shannon, R.D. Revised effective ionic radii and systematic studies of interatomie distances in halides and chaleogenides. Acta Cryst. 1976, 32, 751–767. [Google Scholar] [CrossRef]
- Schlickum, S.; Moghekar, A.; Simpson, J.C.; Steglich, C.; O’Brien, R.J.; Winterpacht, A.; Endele, S.U. LETM1, a gene deleted in Wolf–Hirschhorn syndrome, encodes an evolutionarily conserved mitochondrial protein. Genomics 2004, 83, 254–261. [Google Scholar] [CrossRef]
- Lupo, D.; Vollmer, C.; Deckers, M.; Mick, D.U.; Tews, I.; Sinning, I.; Rehling, P. Mdm38 is a 14-3-3-like receptor and associates with the protein synthesis machinery at the inner mitochondrial membrane. Traffic 2011, 12, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Frazier, A.E.; Taylor, R.D.; Mick, D.U.; Warscheid, B.; Stoepel, N.; Meyer, H.E.; Ryan, M.T.; Guiard, B.; Rehling, P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 2006, 172, 553–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szyrach, G.; Ott, M.; Bonnefoy, N.; Neupert, W.; Herrmann, J.M. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003, 22, 6448–6457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piao, L.; Li, Y.; Kim, S.J.; Byun, H.S.; Huang, S.M.; Hwang, S.K.; Yang, K.J.; Park, K.A.; Won, M.; Hong, J.; et al. Association of LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP production and necrotic cell death. Cancer Res. 2009, 69, 3397–3404. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Zhang, J.; Zhang, X.; Huang, C.; Hu, G.; Li, S.; Xie, T.; Liu, M.; Xu, Y. Suppression of LETM1 by siRNA inhibits cell proliferation and invasion of bladder cancer cells. Oncol. Rep. 2017, 38, 2935–2940. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ni, W.; Cui, C.; Qi, W.; Piao, L.; Xuan, Y. Identification of LETM1 as a marker of cancer stem-like cells and predictor of poor prognosis in esophageal squamous cell carcinoma. Hum. Pathol. 2018, 81, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2018, in press. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Q.-T.; Stathopulos, P.B. Molecular Mechanisms of Leucine Zipper EF-Hand Containing Transmembrane Protein-1 Function in Health and Disease. Int. J. Mol. Sci. 2019, 20, 286. https://doi.org/10.3390/ijms20020286
Lin Q-T, Stathopulos PB. Molecular Mechanisms of Leucine Zipper EF-Hand Containing Transmembrane Protein-1 Function in Health and Disease. International Journal of Molecular Sciences. 2019; 20(2):286. https://doi.org/10.3390/ijms20020286
Chicago/Turabian StyleLin, Qi-Tong, and Peter B. Stathopulos. 2019. "Molecular Mechanisms of Leucine Zipper EF-Hand Containing Transmembrane Protein-1 Function in Health and Disease" International Journal of Molecular Sciences 20, no. 2: 286. https://doi.org/10.3390/ijms20020286