The Universal 3D QSAR Model for Dopamine D2 Receptor Antagonists
Abstract
:1. Introduction
2. Results and Discussion
2.1. Studied Compounds
2.2. Molecular Docking
2.3. Molecular Alignment
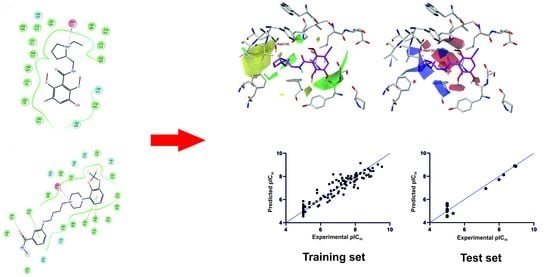
2.4. CoMFA Statistics
2.5. Validation of CoMFA Model
2.6. Contour Map and Its Mapping onto Receptor Structure
3. Materials and Methods
3.1. Selection and Preparation of Compounds
3.2. Molecular Docking
3.3. CoMFA Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMPA | α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid |
| ANN | Artificial neural network |
| CoMFA | Comparative molecular field analysis |
| CoMSiA | Comparative molecular similarity indices analysis |
| ECL | Extracellular loop |
| LOO | Leave-one-out |
| MLR | Multiple linear regressions |
| GPCRs | G protein-coupled receptors |
| ONC | Optimum number of components |
| PLS | Partial least square |
| QSAR | Quantitative structure–activity relationship |
| SEE | Standard error of estimate |
| SP | Standard precision |
| TM | Transmembrane helix |
References
- Stępnicki, P.; Kondej, M.; Kaczor, A.A. Current concepts and treatments of schizophrenia. Molecules 2018, 23, 2087. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.C.; Tsai, S.-J. New targets for schizophrenia treatment beyond the dopamine hypothesis. Int. J. Mol. Sci. 2017, 18, 1689. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Che, T.; Levit, A.; Shoichet, B.K.; Wacker, D.; Roth, B.L. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 2018, 555, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Urniaż, R.D.; Jóźwiak, K. X-ray crystallographic structures as a source of ligand alignment in 3D-QSAR. J. Chem. Inf. Model. 2013, 53, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Kubinyi, H. Strategies and recent technologies in drug discovery. Pharmazie 1995, 50, 647–662. [Google Scholar]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef]
- Clark, R.D.; Abrahamian, E. Using a staged multi-objective optimization approach to find selective pharmacophore models. J. Comput. Aided Msol. Des. 2009, 23, 765–771. [Google Scholar] [CrossRef]
- Kaczor, A.A.; Żuk, J.; Matosiuk, D. Comparative molecular field analysis and molecular dynamics studies of the dopamine D2 receptor antagonists without a protonatable nitrogen atom. Med. Chem. Res. 2018, 27, 1149–1166. [Google Scholar] [CrossRef] [Green Version]
- Kaczor, A.A.; Targowska-Duda, K.M.; Patel, J.Z.; Laitinen, T.; Parkkari, T.; Adams, Y.; Nevalainen, T.J.; Poso, A. Comparative molecular field analysis and molecular dynamics studies of α/β hydrolase domain containing 6 (ABHD6) inhibitors. J. Mol. Model. 2015, 21, 250. [Google Scholar] [CrossRef]
- Fatemi, M.H.; Dorostkar, F. QSAR prediction of D2 receptor antagonistic activity of 6-methoxy benzamides. Eur. J. Med. Chem. 2010, 45, 4856–4862. [Google Scholar] [CrossRef]
- Wang, Q.; Mach, R.H.; Luedtke, R.R.; Reichert, D.E. Subtype selectivity of dopamine receptor ligands: Insights from structure and ligand-based methods. J. Chem. Inf. Model. 2010, 50, 1970–1985. [Google Scholar] [CrossRef] [PubMed]
- Luedtke, R.R.; Mishra, Y.; Wang, Q.; Griffin, S.A.; Bell-Horner, C.; Taylor, M.; Vangveravong, S.; Dillon, G.H.; Huang, R.Q.; Reichert, D.E.; et al. Comparison of the binding and functional properties of two structurally different D2 dopamine receptor subtype selective compounds. ACS Chem. Neurosci. 2012, 3, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Kondej, M.; Wróbel, T.M.; Silva, A.G.; Stępnicki, P.; Koszła, O.; Kędzierska, E.; Bartyzel, A.; Biała, G.; Matosiuk, D.; Loza, M.I.; et al. Synthesis, pharmacological and structural studies of 5-substituted-3-(1-arylmethyl-1,2,3,6-tetrahydropyridin-4-yl)-1H-indoles as multi-target ligands of aminergic GPCRs. Eur. J. Med. Chem. 2019, 180, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Kaczor, A.A.; Silva, A.G.; Loza, M.I.; Kolb, P.; Castro, M.; Poso, A. Structure-Based Virtual Screening for Dopamine D2 Receptor Ligands as Potential Antipsychotics. ChemMedChem 2016, 11, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Kaczor, A.A.; Targowska-Duda, K.M.; Budzyńska, B.; Biała, G.; Silva, A.G.; Castro, M. In vitro, molecular modeling and behavioral studies of 3-{[4-(5-methoxy-1H-indol-3-yl)-1,2,3,6-tetrahydropyridin-1-yl]methyl}-1,2-dihydroquinolin-2-one (D2AAK1) as a potential antipsychotic. Neurochem. Int. 2016, 96, 84–99. [Google Scholar] [CrossRef]
- Kondej, M.; Bartyzel, A.; Pitucha, M.; Wróbel, T.M.; Silva, A.G.; Matosiuk, D.; Castro, M.; Kaczor, A.A. Synthesis, Structural and Thermal Studies of 3-(1-Benzyl-1,2,3,6-tetrahydropyridin-4-yl)-5-ethoxy-1H-indole (D2AAK1_3) as Dopamine D₂ Receptor Ligand. Molecules 2018, 23, 2249. [Google Scholar] [CrossRef]
- Hevener, K.E.; Ball, D.M.; Buolamwini, J.K.; Lee, R.E. Quantitative structure-activity relationship studies on nitrofuranyl anti-tubercular agents. Bioorg. Med. Chem. 2008, 16, 8042–8053. [Google Scholar] [CrossRef]
- Clark, R.D.; Fox, P.C. Statistical variation in progressive scrambling. J. Comput. Aided Mol. Des. 2004, 18, 563–576. [Google Scholar] [CrossRef]
- Yuan, S.; Vogel, H.; Filipek, S. The role of water and sodium ions in the activation of the μ-opioid receptor. Angew. Chem. 2013, 52, 10112–10115. [Google Scholar] [CrossRef] [PubMed]
- Bartuzi, D.; Kaczor, A.A.; Matosiuk, D. Interplay between Two Allosteric Sites and Their Influence on Agonist Binding in Human μ Opioid Receptor. J. Chem. Inf. Model. 2016, 56, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Livingston, K.E.; Stanczyk, M.A.; Burford, N.T.; Alt, A.; Canals, M.; Traynor, J.R. Pharmacologic Evidence for a Putative Conserved Allosteric Site on Opioid Receptors. Mol. Pharmacol. 2018, 93, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Hothersall, J.D.; Torella, R.; Humphreys, S.; Hooley, M.; Brown, A.; McMurray, G.; Nickolls, S.A. Residues W320 and Y328 within the binding site of the μ-opioid receptor influence opiate ligand bias. Neuropharmacology 2017, 118, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Bock, A.; Merten, N.; Schrage, R.; Dallanoce, C.; Bätz, J.; Klöckner, J.; Schmitz, J.; Matera, C.; Simon, K.; Kebig, A.; et al. The allosteric vestibule of a seven transmembrane helical receptor controls G-protein coupling. Nat. Commun. 2012, 3, 1044. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2018-2: LigPrep, Schrödinger; LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-2: Epik, Schrödinger; LLC: New York, NY, USA, 2018.
- Schrödinger Release 2018-2: Glide, Schrödinger; LLC: New York, NY, USA, 2018.



| Components | Q2 | cSDEP | dQ2/dR2yy |
|---|---|---|---|
| 2 | 0.54 | 0.95 | 0.91 |
| 3 | 0.61 | 0.88 | 0.94 |
| 4 | 0.62 | 0.86 | 1.14 |
| 5 | 0.61 | 0.87 | 1.19 |
| 6 | 0.59 | 0.89 | 1.30 |
| 7 | 0.57 | 0.93 | 1.32 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zięba, A.; Żuk, J.; Bartuzi, D.; Matosiuk, D.; Poso, A.; Kaczor, A.A. The Universal 3D QSAR Model for Dopamine D2 Receptor Antagonists. Int. J. Mol. Sci. 2019, 20, 4555. https://doi.org/10.3390/ijms20184555
Zięba A, Żuk J, Bartuzi D, Matosiuk D, Poso A, Kaczor AA. The Universal 3D QSAR Model for Dopamine D2 Receptor Antagonists. International Journal of Molecular Sciences. 2019; 20(18):4555. https://doi.org/10.3390/ijms20184555
Chicago/Turabian StyleZięba, Agata, Justyna Żuk, Damian Bartuzi, Dariusz Matosiuk, Antti Poso, and Agnieszka A. Kaczor. 2019. "The Universal 3D QSAR Model for Dopamine D2 Receptor Antagonists" International Journal of Molecular Sciences 20, no. 18: 4555. https://doi.org/10.3390/ijms20184555